Scope & Usage
Scope
SMAP snp-seq designs HiPlex primers encompassing dedicated polymorphic SNP sites, while avoiding neighboring ‘background’ SNPs. It is a simple application to design primer panels for targeted multiplex amplicon resequencing taking known polymorphisms into account, and can be directed to pre-selected locations like GBS loci or candidate genes.
Input
SMAP snp-seq only requires a reference sequence FASTA file and one VCF file with the SNPs that need to be targeted. Optionally, one may provide a BED file with selected template regions, or a VCF file with ‘background’ SNPs that need to be avoided during primer design. Last, one may create a customized reference for a particular sample set by providing a VCF file with SNPs where the reference nucleotide is substituted by the alternative nucleotide in the reference sequence prior to primer design.
Output
Integration in the SMAP workflow
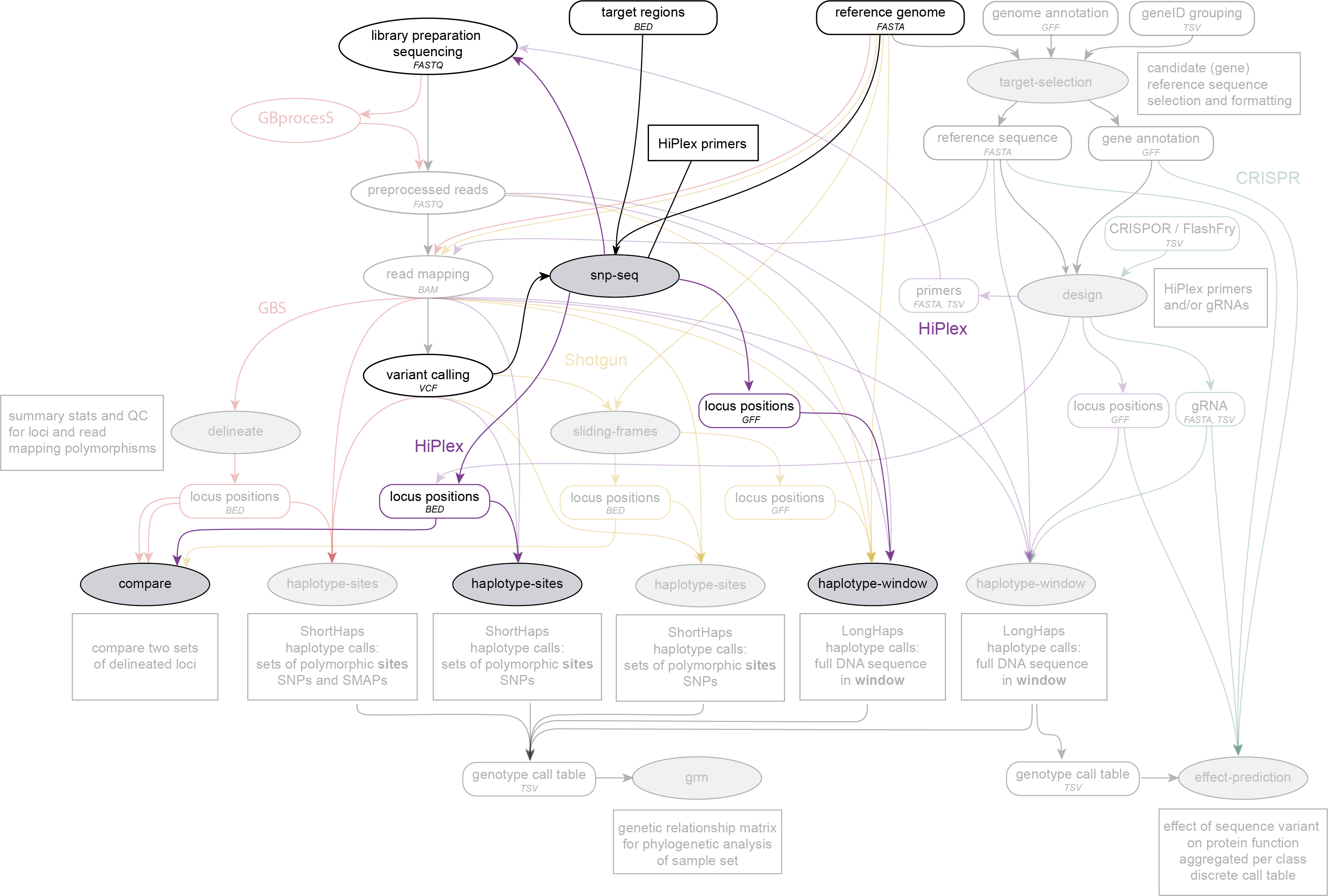
SMAP snp-seq is run on a reference sequence FASTA file and one or two VCF files, after variant calling and before SMAP haplotype-sites or SMAP haplotype-window. SMAP snp-seq designs primer panels for HiPlex amplicon sequencing.
Guidelines for variant calling
See Veeckman et al. (2019) for a comparison of different SNP calling methods.
Commands & options
Mandatory options for SMAP snp-seq
SMAP snp-seq only needs a reference sequence and known SNP positions to target.
--reference ### The FASTA file with the reference sequence [no default].--target_vcf ## The VCF file with SNPs [no default].Command line options
See tabs below for command line options and specific filter options.
Input data options:
-i,--input_directory##### (str) ## Input directory [current directory].--template_region######## (str) ## Name of the BED file in the input directory containing the genomic coordinates of regions wherein primers must be designed [no BED file provided].--background_vcf############## Name of the VCF file in the input directory containing target SNPs [no VCF file with target SNPs provided].--customized_reference########## Name of the VCF file in the input directory containing non-polymorphic differences between the reference genome sequence and the samples for primer design [no VCF file with reference genome differences provided].
Amplicon design options:
--maximum_variant_distance####### (int) ### Maximum distance (in bp) between two variants to be included in the same template region [500].--flanking_region########## (int) ### Length of the flanking region (in bp) to be added on both ends of the central template region [half of the maximum variant distance].--maximum_target_size############### (int) ### Maximum size (in bp) of a target region [10].--minimum_target_distance############ (int) ### Minimum distance (in bp) between two target regions in a template [0].--minimum_amplicon_size####### (int) ### Minimum size of an amplicon (incl. primers) in bp [100].--maximum_amplicon_size####### (int) ### Maximum size of an amplicon (incl. primers) in bp [110].--offset####################### (int) ### Size of the offset at the 5’ and 3’ end of each target region. Variants in the region covered by offset are not tagged as targets for primer design [0, all variants are potential targets].--minimum_primer_size######## (int) ### Minimum size (in bp) of a primer [18].--maximum_primer_size######## (int) ### Maximum size (in bp) of a primer [27].--optimal_primer_size######## (int) ### Optimal size (in bp) of a primer [20].--maximum_mispriming###### (int) ### Maximum allowed weighted similarity of a primer with the same template and other templates [12].--maximum_number_degenerate_nucleotides## (int) ### Maximum number of degenerate nucleotides (N) in a primer sequence [0].--region_extension########### (int) ### Extend template regions in the BED file provided via the--template_regionoption at their 5’ end 3’ end with the provided value [0, no template region extension].--retain_overlap############# Retain overlap among template regions [overlap in template regions is removed].--split_template_region######### Split the regions in the BED file provided via the--template_regionoption in multiple templates based on the maximum_variant_distance [template regions are not split].
Options may be given in any order.
Command to run SMAP snp-seq:
python3 SMAP_snp-seq.py -i /path/to/dir/ --target_vcf variants.vcf --reference genome.fasta
-o,--output_directory### (str) ### Path to the output directory [current directory].--border_length##### (int) ### Border size used in the GFF file that defines the windows for SMAP haplotype-window [10].--suffix########## (str) ### Suffix added to output files [set_1].
Options may be given in any order.
Command to run SMAP snp-seq with adjusted border length and suffix to denote the design settings:
python3 SMAP_snp-seq.py -i /path/to/dir/ --target_vcf variants.vcf --reference genome.fasta --border_length 10 --suffix Lp_120_180bp
Example commands
Basic command to run SMAP snp-seq with target SNPs:
python3 SMAP_snp-seq.py -i /path/to/dir/ --target_vcf variants.vcf --reference genome.fasta
Command to run SMAP snp-seq for a set of target SNPs while avoiding background SNPs for primer design:
python3 SMAP_snp-seq.py -i /path/to/dir/ --target_vcf variants.vcf --reference genome.fasta --background_vcf background_snps.vcf
Command to run SMAP snp-seq with a set of SNPs to substitute in a customized reference sequence:
python3 SMAP_snp-seq.py -i /path/to/dir/ --target_vcf variants.vcf --reference genome.fasta --customized_reference reference_variants.vcf
Command to run SMAP snp-seq for a specific set of loci (template regions):
python3 SMAP_snp-seq.py -i /path/to/dir/ --target_vcf variants.vcf --reference genome.fasta --template_region gbs_centralregion.bed