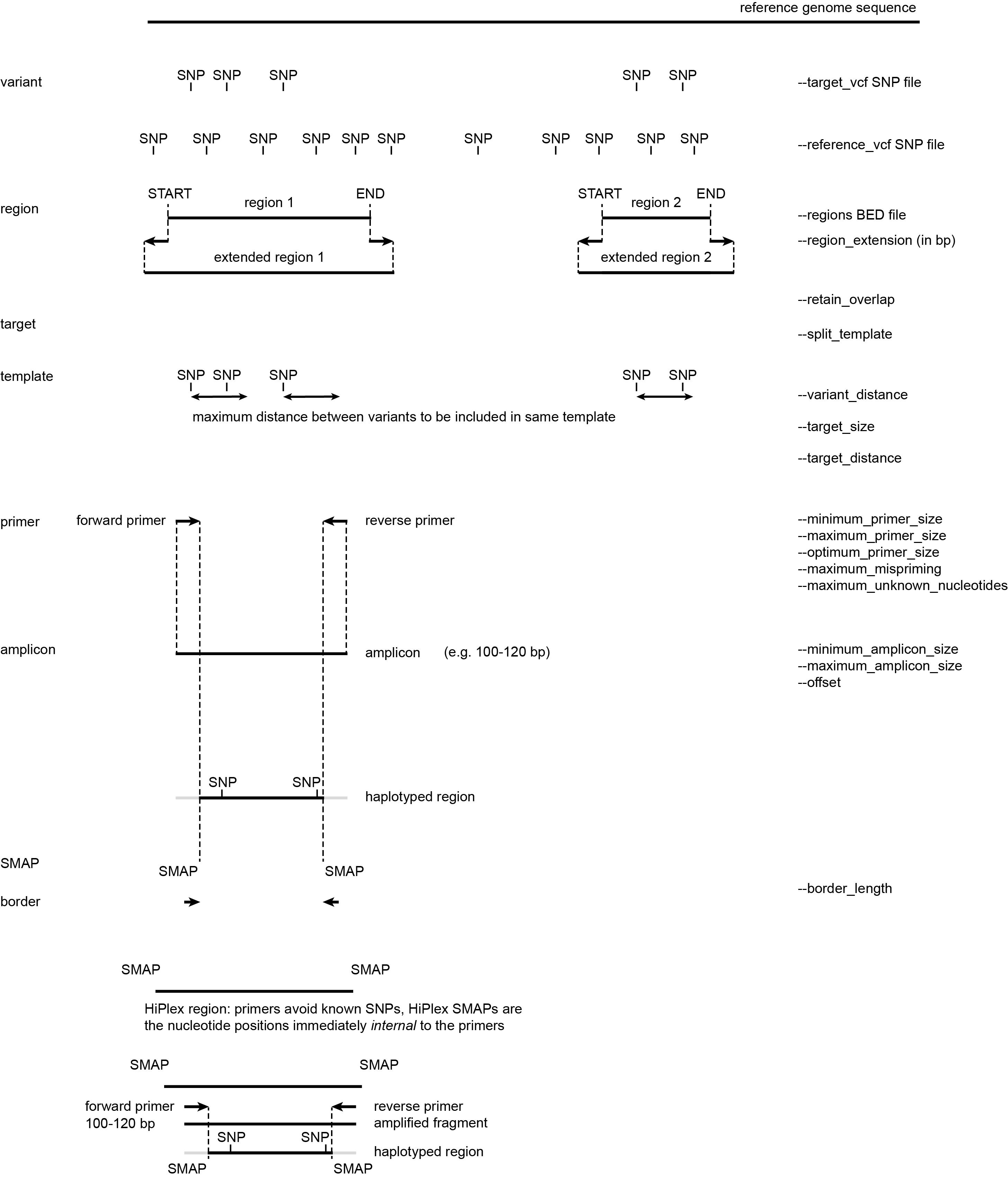Several applications of molecular markers iterate between marker discovery (e.g. WGS or GBS) and targeted screening by HiPlex amplicon sequencing (e.g. SNP-seq).
The SMAP snp-seq module fills the gap between those strategies: it takes SNP variants identified in a large screen, and allows to automatically design primers flanking selected SNPs or within selected regions, in both cases avoiding all known SNPs at primer binding sites.
Several input files are optionally provided to define the SNPs and/or regions to be targeted, and the SNPs to avoid during primer design.
SMAP snp-seq also generates the coordinate files for downstream analysis of HiPlex read data: a BED file with SMAPs for downstream analysis with SMAP haplotype-sites, or a GFF file with border positions for SMAP haplotype-window.
In addition, several parameters can be set to define distances between SNPs and/or loci.
In principle, it is possible to
a priori define regions to be targeted (such as 1 kb regions at 1 Mb intervals) to design a HiPlex set that covers the entire genome at a fixed marker distance (for an example in potato, see
de la O Leyva-Pérez et al. (2022)).
In addition, SMAP snp-seq can also be used to transfer GBS marker sets to HiPlex marker sets, by providing the ‘CentralRegions’ BED file generated by SMAP delineate, and a VCF file generated with e.g. GATK, while specifying minimum and maximum amplicon size for the designed HiPlex fragments (see scheme below).
Defining regions according to different scenario’s
Schematic overview of design steps
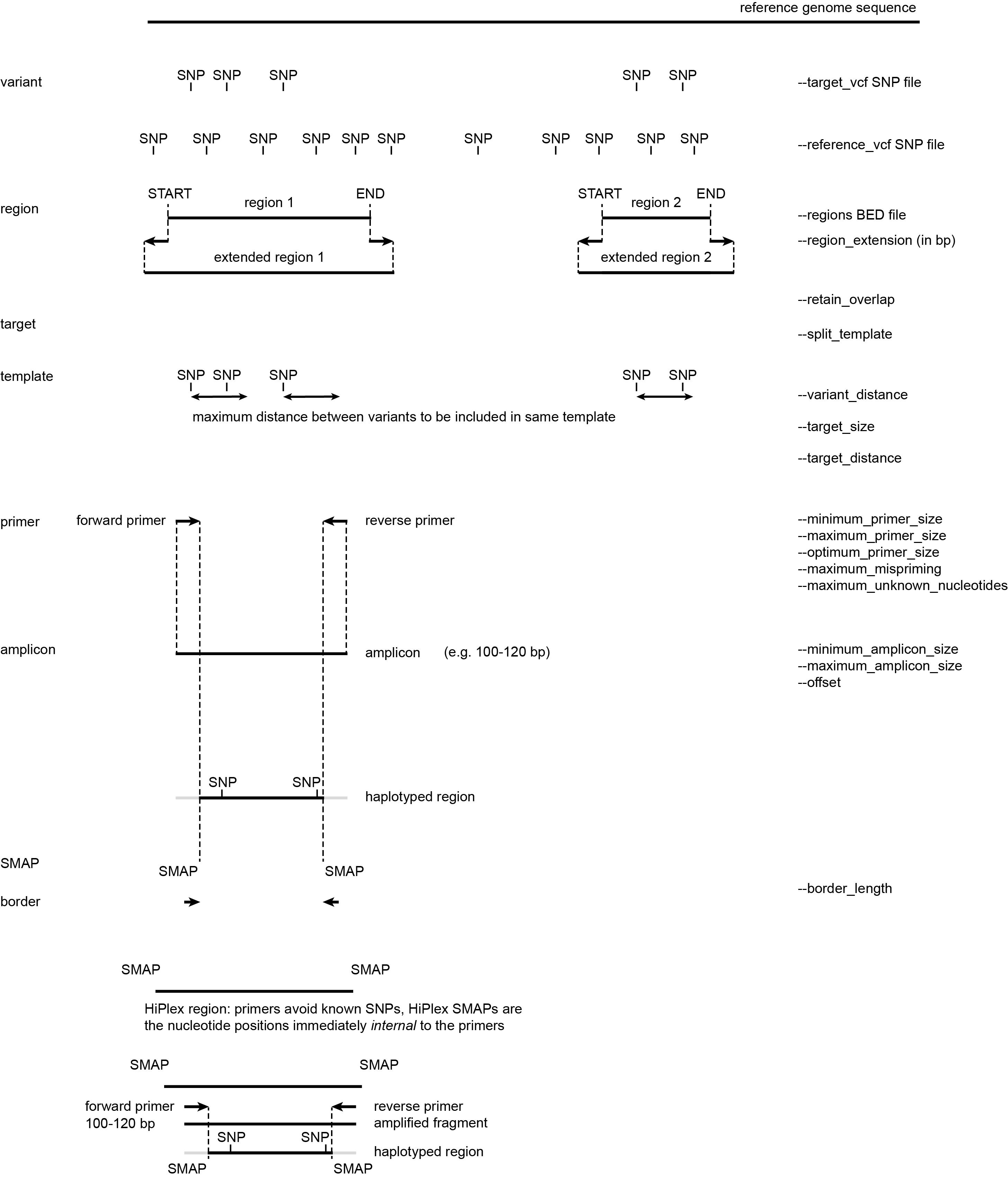
The different options applied by SMAP snp-seq depend on the type of data with which SNPs were obtained in previous steps.
These are illustrated by a simplified drawing of whole genome shotgun (WGS) sequencing data and/or genotyping-by-sequencing (GBS) data (Figure 1), but SNPs from other sequencing library types (e.g. RNA-seq data, probe capture data) can be used as input as well.

Graphical representation of sequencing reads (grey bars) containing SNPs (yellow squares) from WGS libraries (upper) or GBS libraries (lower) that are mapped onto a reference genome sequence.
These representations will be used to demonstrate the different options of SMAP snp-seq.
HIW: Extending regions with a (small) number of nucleotides can be advantageous for primer design, because the extended parts may provide more possibilities for primer3 to design primers around targets. Extending regions might be interesting for regions with a low SNP density, as it is unlikely that unknown SNPs are located directly flanking the region.