SMAP sliding frame
This is the manual for the complementary SMAP utility tool SMAP sliding-frames of the SMAP package.
The first step prior to running SMAP haplotype-sites is the definition of the locus start and end points.
The python script SMAP_sliding-frame.py should be used to define sliding frames covering SNPs and/or structural variants in Shotgun data (currently provided as Python3 script in the SMAP utility tools).
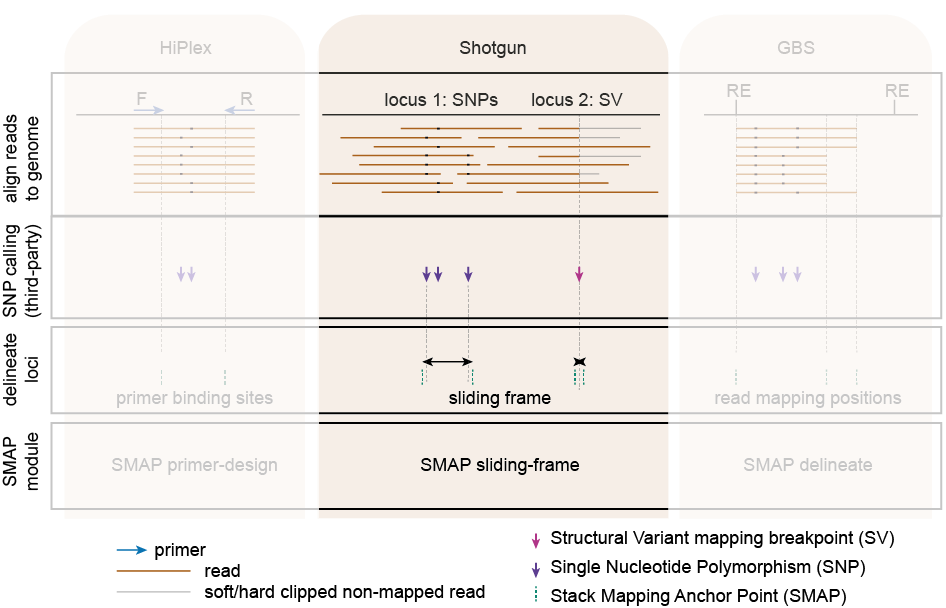
The module SMAP delineate should be run for GBS data to define relevant loci and read mapping polymorphisms in a data-driven manner.
A module called SMAP design will be launched in the near future for integrated design of HiPlex PCR primers and downstream analysis with SMAP haplotype-sites and/or SMAP haplotype-window.