SMAP haplotype-sites
This is the manual for the SMAP haplotype-sites component of the SMAP package.
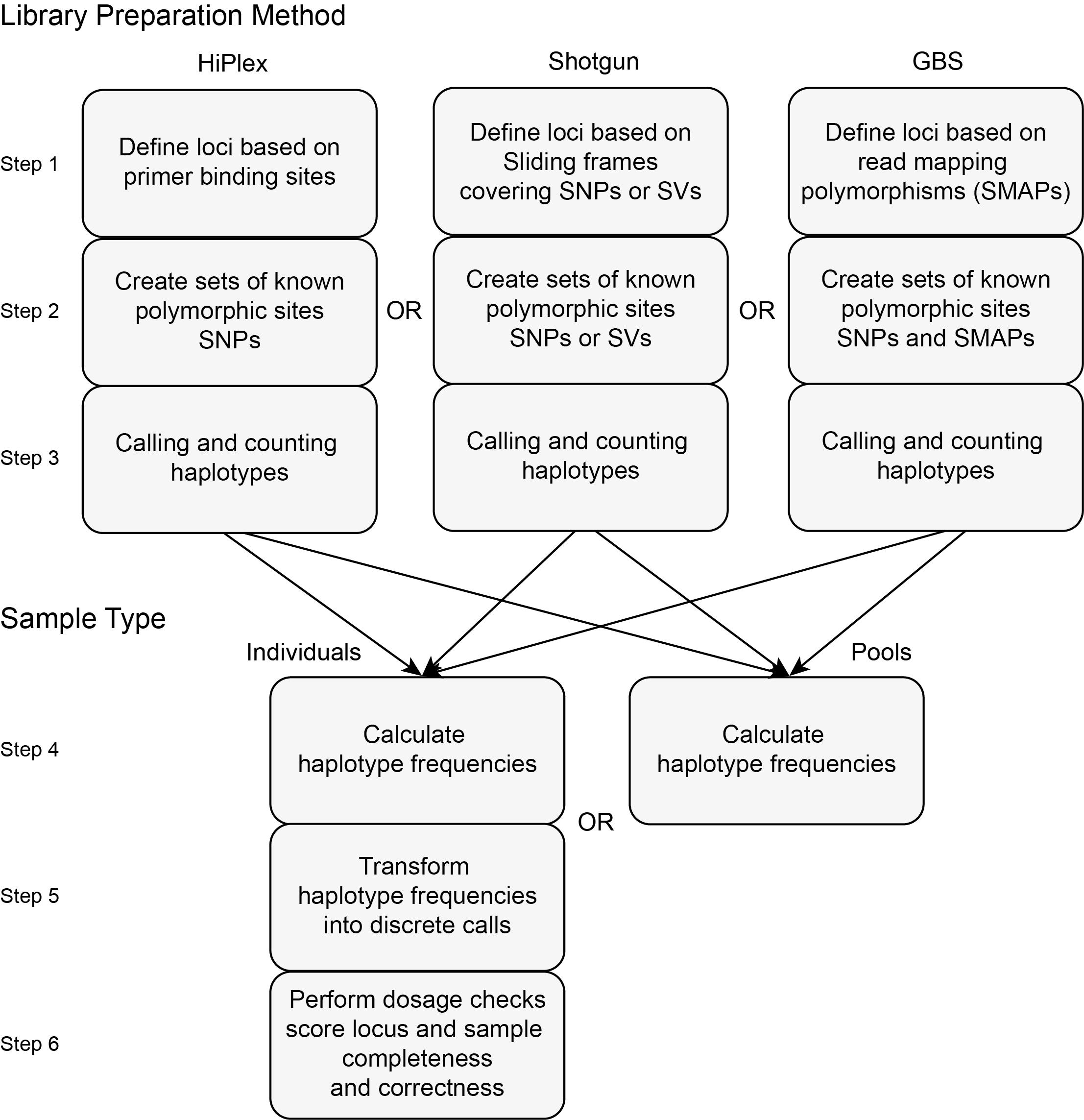
The first step prior to running SMAP haplotype-sites is the definition of the locus start and end points.
For HiPlex and Shotgun data these can be created using Python scripts provided in the Utility tools.
For GBS data, SMAP delineate should be run to define relevant loci and read mapping polymorphisms in a data-driven manner.
The scheme below depicts the two major distinctions, concerning library preparation method and sample type, implemented in this program.