Scope & Usage
Scope
It’s main functions are to:
Filter for haplotypes with edits in a defined region of interest (ROI; e.g. surrounding the PAM site for CRISPR/Cas experiments) to eliminate noise from the genotype table.
Substitute the segment of the original reference gene sequence by the observed haplotype, keeping track of all relevant coordinates of intron-exon borders, translational start and stop codons, and the open reading frame (ORF), and predict the resulting (mutated) protein sequence.
Compare the novel predicted protein sequence to the original reference protein and estimate the fraction of the protein length that is still encoded by the novel (mutated) allele.
Use a threshold for the %protein length required for (partial) loss of function, and classify all haplotypes by effect class (no/minimal effect, intermediate effect, loss-of-function).
Aggregate all observed haplotypes and sum their relative frequencies per effect class.
Finally, discretize the genotype calls as homozygous or heterozygous for reference versus loss of function at a user defined minimal effect level.
Plot summary statistics of editing “fingerprints” across the data set to allow the user to optimize parameter setting accoring to their experimental data.
Within the SMAP package, the modules SMAP target-selection, SMAP design, SMAP haplotype-window and SMAP effect-prediction are designed to provide a seamless workflow from target selection (e.g. candidate genes), integrated primer and gRNA design, multiplex resequencing of target loci across large plant collections, followed by identification of all observed haplotypes (naturally occuring or CRISPR-induced sequence variants), the prediction of functional effects of sequence variants at the protein level to identify (partial) loss-of-function (LOF) alleles, and finally aggregate and discretize genotype calls in an integrated genotype table with the homozygous/heterozygous presence of LOF alleles per locus per sample. The overarching goal of this entire workflow is to identify carriers of LOF alleles for functional analysis, or for genotype-phenotype associations.
Specifically, the underlying concepts of SMAP effect-prediction exploit:
Modularity, compatibility throughout the entire workflow.
Flexibility in design (scalability to complex multi-amplicon / multi-gRNA design per gene).
Predicted effect of the observed mutation on the encoded protein level.
Customized aggregation of effects per haplotype (thresholds).
Customized aggregation of alleles per effect class per locus (thresholds).
Discretizing the complex haplotype table to a simple homozygous / heterozygous LOF effect per locus per sample.
Single command line operation per module.
Traceable output (discrete LOF-call genotype table, alignments, VCF-encoded variants, predicted proteins).
Biology-driven decisions.
Integration in the SMAP workflow
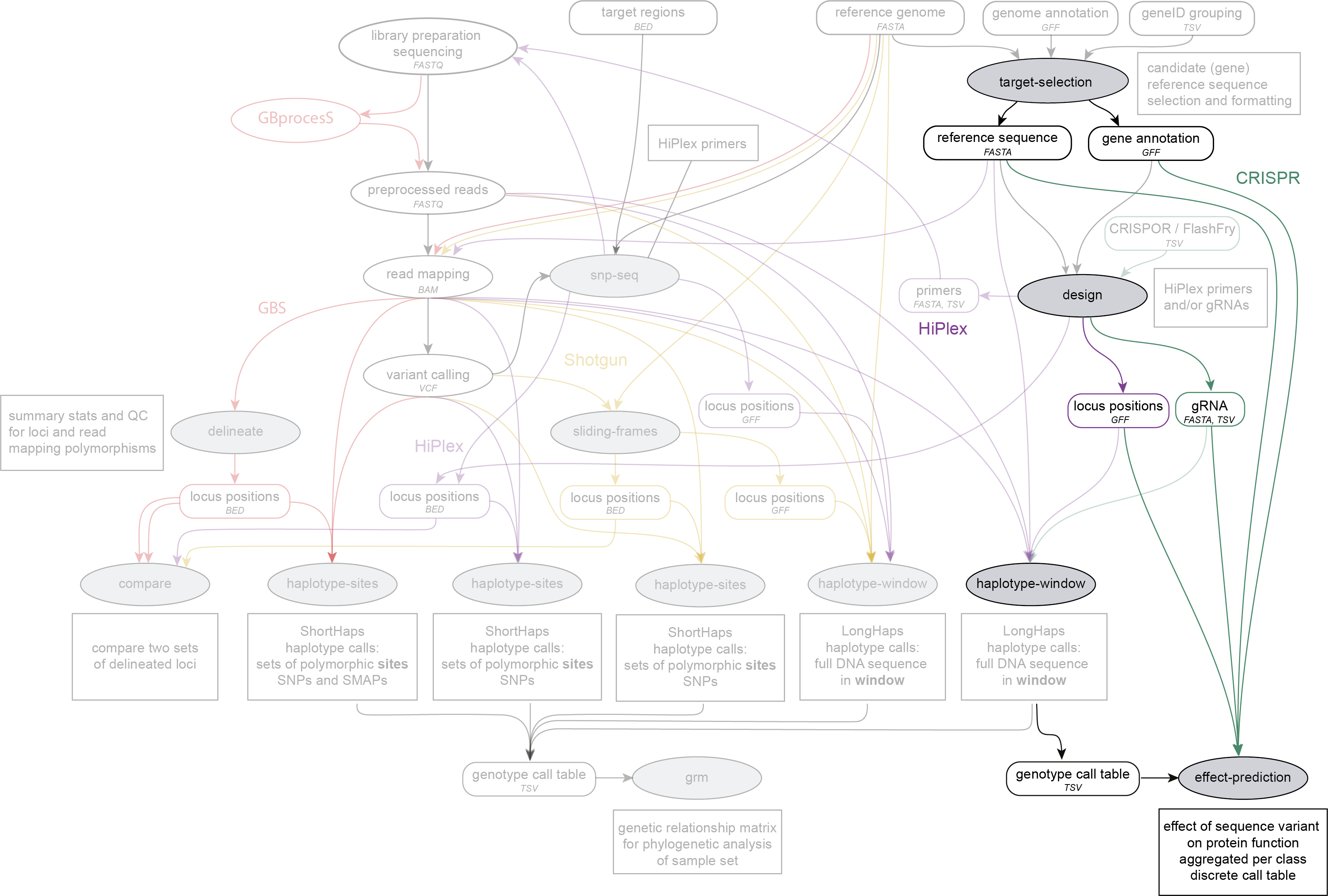
SMAP effect-prediction is run on a reference sequence FASTA file with candidate genes, and associated GFF file with gene annotations created by SMAP target-selection, optionally a gRNA FASTA file and locus positions created by SMAP design, and a genotype call table created by SMAP haplotype-window. SMAP effect-prediction works on HiPlex data.
Guidelines for SMAP effect-prediction
These tabs provide a decision scheme to guide you to the correct parameter settings.
option -u.option -r and upper bounds option -s around the cutsite, as nucleotide distance.option -f relative to the gRNA 5’ endoption -p.-r and upper bounds -s, offset -f or -p, and ROI for gRNAs located on the forward and/or reverse strand -u.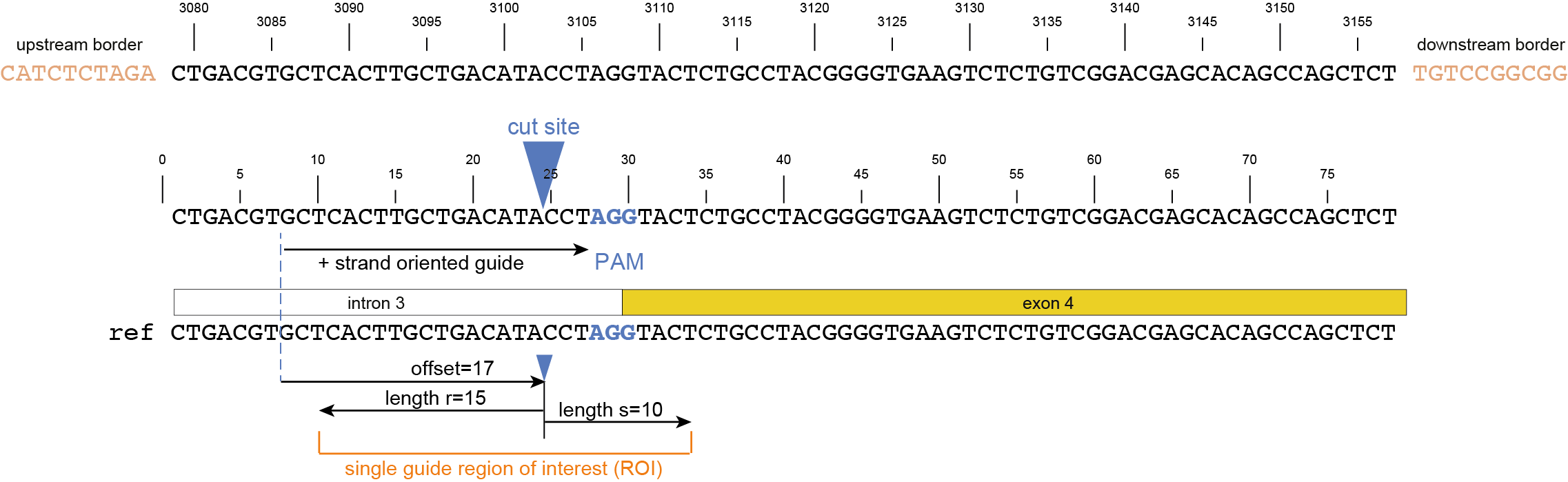
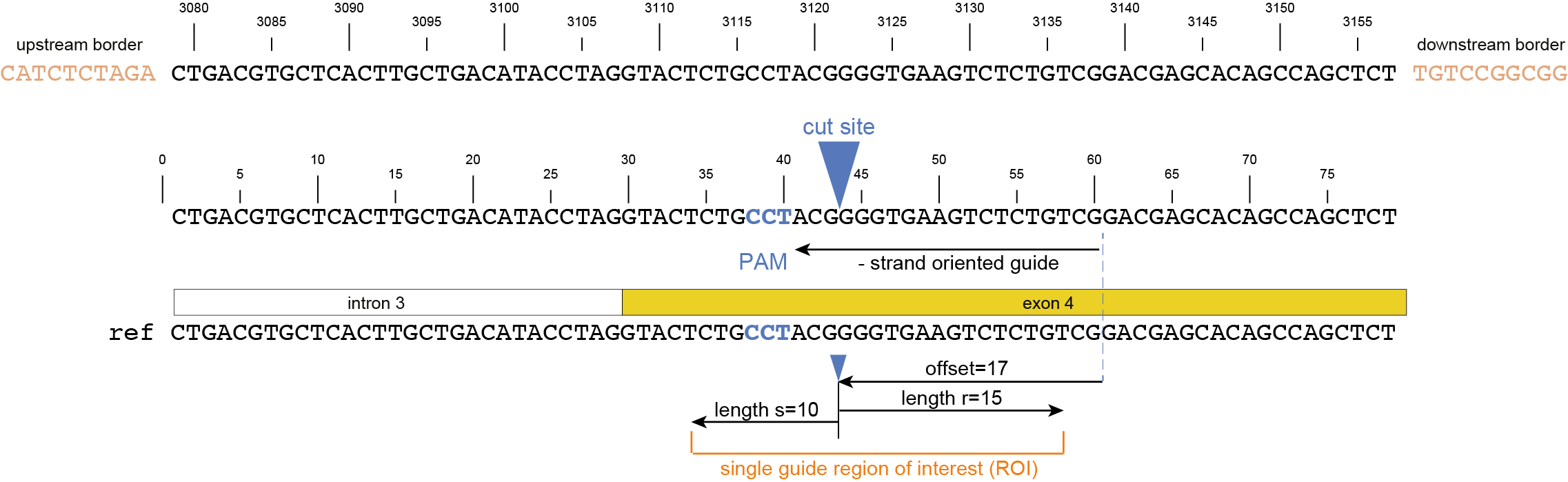
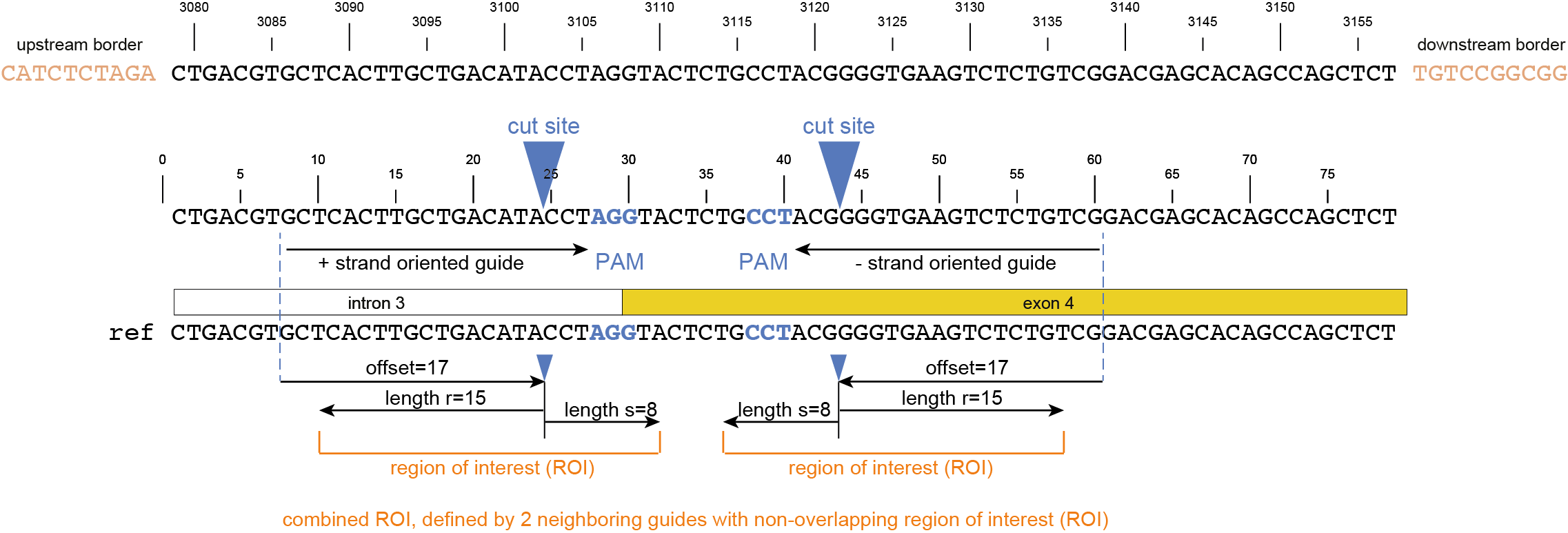
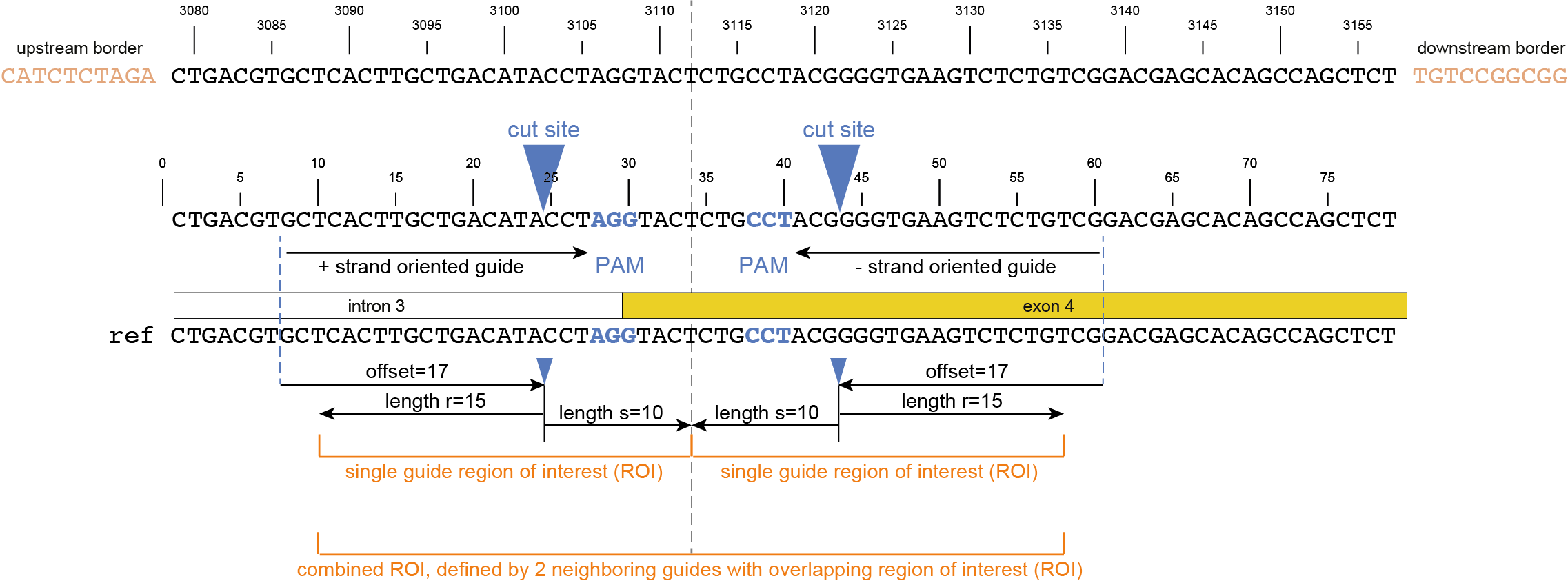
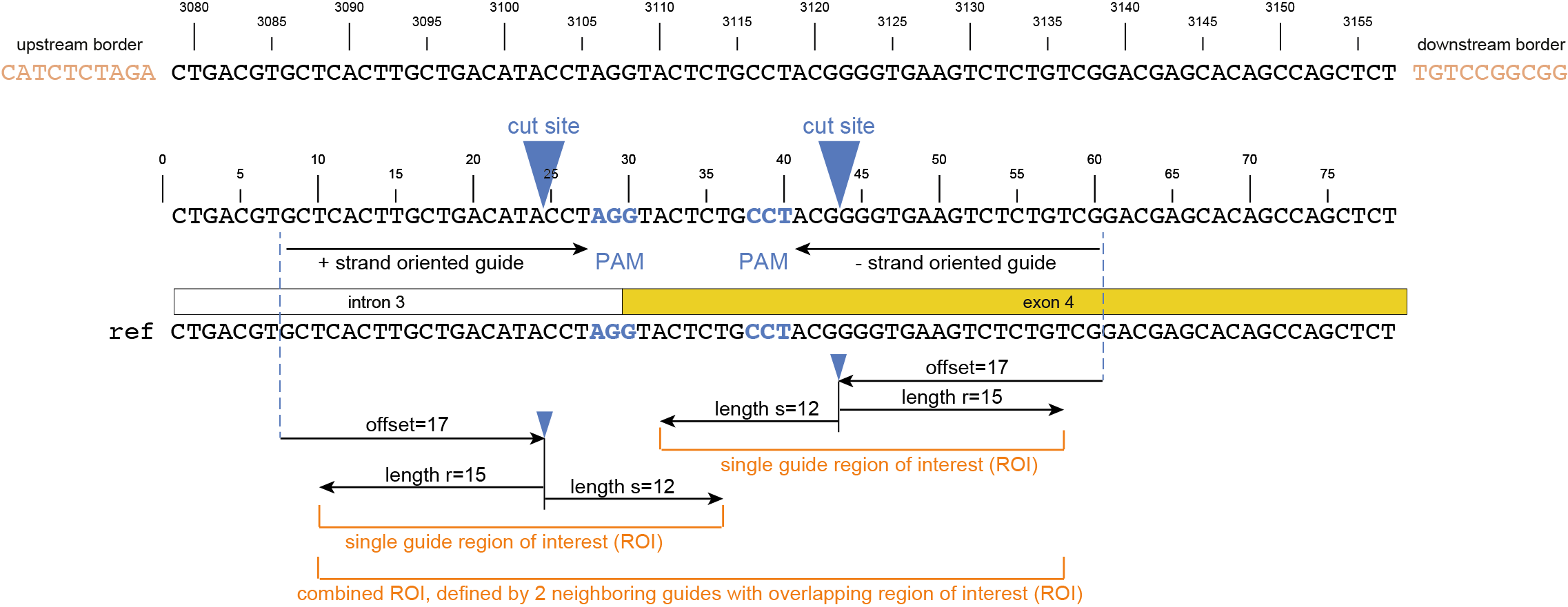
Do you want to predict the effect of mutations in the ROI on the encoded protein?
option -a. CDS features must be located on the positive strand.option -i to transform frequency data of haplotypes into discrete genotype calls (homozygous reference, heterozygous, homozygous mutated at the predicted protein effect class (minor, major effect)).option -e to get binary presence/absence data.--disable_protein_prediction.Do you want to predict the effect of haplotype mutations on the encoded protein?
option -a. CDS features must be located on the positive strand.Do you want to aggregate the haplotype frequencies based on their effect on the encoded protein?
Do you want to discretize the aggregated frequencies into discrete calls?
option -i to transform frequency data of haplotypes into discrete genotype calls (homozygous reference, heterozygous, homozygous mutated at the predicted protein effect class (minor, major effect)).option -e to get binary presence/absence data.--disable_protein_prediction.Commands & options
Mandatory options for SMAP effect-prediction
It is mandatory to specify the files with the haplotype frequency table, the associated reference sequence, the set of gRNA sequences and GFF with positional information of CDS.
- Input and output information
It is mandatory to specify the files with the haplotype frequency table, the associated reference sequence, the set of gRNA sequences, and a GFF3 with structural gene annotation. First, the haplotype frequency table should be generated using SMAP haplotype-window. Second, the same reference sequence that was used to generate the haplotype frequency table with SMAP haplotype-window must be provided to SMAP effect-prediction. Third, haplotype calling occurred within a ‘window’, defined by two borders (typically the 10 nucleotides at the 3’ of the HiPlex primers). The position of the windows are provided to SMAP effect-prediction by a GFF3 file containing the positions of these borders. A single gff entry corresponds to one border, and two borders must be linked together to form a window by using a shared NAME attribute value. All borders must be specified in the ‘+’ orientation to the reference genome. Finally, a GFF3 file defining the gene and CDS information should be provided. For your convenience, all these input files can be prepared with the modules SMAP target-selection and SMAP design.
Regarding input files, there is only one file that is considered optional: a GFF3 file of the gRNA positions. These gRNA positions allow SMAP effect-prediction to filter haplotypes to collapse those haplotypes that only contain variations outside a user-defined range around the cut site defined by the gRNA where ‘true positive’ variation resulting from CRISPR/Cas activity is expected to occur. Each gRNA should be a single gff entry, with a ‘+’ or ‘-’ orientation compared to the reference. Additionally, each gRNA should have a unique NAME attribute that specifies its target locus.
The locations of the gRNAs are not enough to specify where the Cas enzyme cuts the DNA for editing. The type of Cas protein used for the editing experiment also determines the offset relative to the position of the gRNA. Therefore, options are available to specify this offset by either using a predefined offset by using the name of the Cas9 protein, or by using a custom offset (i.e. number of nucleotides).
-u, --gRNAs ############### (str) ### .gff file containing the gRNA coordinates, must contain NAME=<> in column 9.-g, --no_gRNA_relative_naming ## (str) ### Change the haplotype naming according to the gRNA coordinates.-p, --cas_protein ########### (str) ### Name of the nuclease used in the experiment. Used to select a predefined offset [CAS9].-f CAS_OFFSET, --cas_offset #### (int) ### Cas offset in number of nucleotides.-s, --cut_site_range_upper_bound ## (str) ### Upper bound for selecting variations from a range around the cut site. Defined in the direction from the cut site towards the PAM.-r, --cut_site_range_lower_bound ## (str) ### Lower bound for selecting variations from a range around the cut site. Defined in the direction from the cut site towards the start of the gRNA binding site.-c, --cpu ### (str) ### Maximum number of allowed processes.Alignment parameters: The default settings below have been determined empirically. As SMAP effect-prediction relies heavily on the alignment of haplotypes to the reference sequence, caution is advised when changing these defaults. For more information on the alignment implementation, we refer to the biopython documentation. Define the parameters to align the haplotype sequences to the reference sequence.
--match_score ##### (str) ###--mismatch_penalty ## (str) ###--gap_open_penalty ## (str) ###--gap_extension #### (str) ###Use thresholds to transform haplotype frequencies into discrete calls using fixed intervals. The assigned intervals are indicated by a running integer. This is only informative for individual samples and not for Pool-Seq data.
-e, --discrete_calls {dominant,dosage} ########## (str) ### Set to “dominant” to transform haplotype frequency values into presence(1)/absence(0) calls per allele, or “dosage” to indicate the allele copy number.-i, --frequency_interval_bounds ################# (str) ### Frequency interval bounds for transforming haplotype frequencies into discrete calls. Custom thresholds can be defined by passing one or more space-separated values (relative frequencies in percentage). For dominant calling, one value should be specified. For dosage calling, an even total number of four or more thresholds should be specified. Default values are invoked by passing either “diploid” or “tetraploid”. The default value for dominant calling (see discrete_calls argument) is 10, both for “diploid” and “tetraploid”. For dosage calling, the default for diploids is “10, 10, 90, 90” and for tetraploids “12.5, 12.5, 37.5, 37.5, 62.5, 62.5, 87.5, 87.5”.--disable_protein_prediction ### (str) ### Disable the estimation of the protein from the haplotypes sequences. All variations within range (-s and -r) of the cut site will be considered as relevant sequence variant with an effect. This option requires --gRNAs.-t, --effect_threshold ####### (str) ### Threshold to determine whether a protein is affected by the haplotype variant sequence or not. For each haplotype, a protein identity score is calculated compared to the reference. Haplotypes for which the protein identity is below the effect threshold, will be marked as encoding an affected protein.For instance, a protein with 10% identity to the reference, is below an effect threshold of 50%, and will be marked as loss-of-function (LOF).Example commands
Example command line to run SMAP effect-prediction with adjusted aggregation thresholds:
smap effect-prediction haplotype-window_genotype_table.tsv genome.fasta borders.gff local_gff_file.gff3 -u gRNAs.gff -p CAS9 -s 10 -r 20 -e dosage -i diploid -t 90
Output
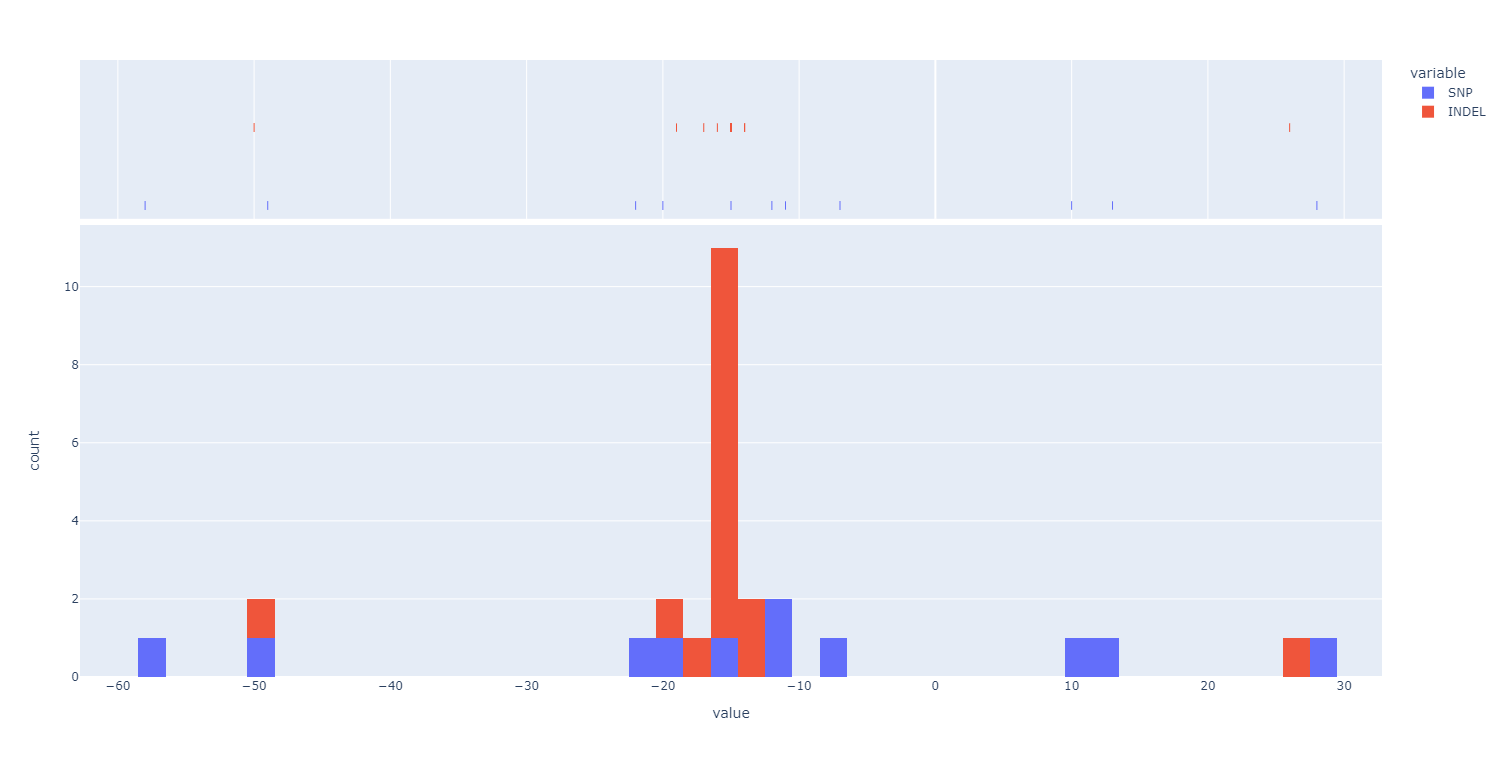
SMAP effect-prediction creates two pre-aggregation tables: annotate.tsv and collapsed.tsv.SMAP effect-prediction creates two post-aggregation tables: aggregated.tsv and discretized.tsv.The following tabs show real experimental data of nine loci. All detected haplotypes are reported using the default settings, demonstrating how annotation and aggregation compresses the genotype call table, and discretization simplifies the calls to heterozygous/homozygous knock-out genotype calls.
Reference |
Locus |
Haplotypes |
target |
edit |
start |
end |
SNP |
INDEL |
Alignment |
FILTER_gRNA_INDEL |
FILTER_gRNA_SNP |
FILTER_gRNA |
Haplotype_Name |
Expected cut site |
atgCheck |
splicingSiteCheck |
stopCodonCheck |
protein_sequence |
pairwiseProteinIdentity (%) |
Effect |
sample1 |
sample2 |
sample3 |
sample4 |
sample5 |
sample6 |
sample7 |
sample8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
gene1 |
gene1_1 |
GGCTCTGTTCTTTTACTCGGCCCTGTTTGACGCTCTGGACACGACCACTCCAAGAGACAGCAACCAGAGGATGCT |
gene1 |
ref |
1790 |
1865 |
nan |
nan |
nan |
nan |
nan |
nan |
ref |
1817 |
MGSSYDPYPSPGADDLFLYLSDLGPASPSAYLDLPPTPQPQPYPQSQQQQQGSKGPTQDMLLPYISSMLMEDDIDDTFFYDYPDNPALLQAQQSFLDILSDDASSPTTTTGTTNSSASVNHSSSDASASAPPTPAAVDSYSPAPAVQFDGFDLDPAAFFSNGANSDLMSSAFLKGMEEANKFLPSQDKLVIDLDPPDDTKRFVLPTRAAENLAPGFNAAATTVPAAVAMAVKEEEVILAALDAALGSGGVVLGRGRRNRLDDDEEDLELQRRSTKQSALQGDGDERDVFEKYIMTCPETCTEQMQQLRIAMQEEAAKEAAVAAGNGKAKGRRGGREVVDLRTLLVHCAQAVASDDRRSATELLRQIKQHASPQGDATQRLAHCFAEGLQARLAGTGSMVYQSLMAKRTSAADILQAYQLYMAAICFKRVVFVFSNNTIYNAALGKMKIHIVDYGIHYGFQWPCFLRWIADREGGPPEVRITGIDLPQPGFRPTQRIEETGRRLSKYAQQFGVPFKYQAIAASKMESIRAEDLNLDPEEVLIVNCLYQFKNLMDESVVIESPRDIVLNNIRKMRPHTFIHAIVNGSFSAPFFVTRFREALFFYSALFDALDTTTPRDSNQRMLIEENLFGRAALNVIACEGTDRVERPETYKQWQVRNQRAGLKQQPLNPDVVQQDIGTSNQEEICDASMS* |
NA |
False |
100.0 |
100.0 |
100.0 |
100.0 |
100.0 |
100.0 |
100.0 |
100.0 |
|||
gene2 |
gene2_1 |
AGTGCTAGGTGATGCTGCGCGAGTACTGCGAGATCTAATCACTCAAGTGGAATCTCTCAGGCAGGAACAATCTGCTCTTG |
gene2 |
ref |
3751 |
3831 |
nan |
nan |
nan |
nan |
nan |
nan |
ref |
3795 |
EFPCRWGRRRRHVSPRYRLGHTSVRVGGRHTLAAPACVCSPPTLPALALHFPWQSSLVPDFLPERIRPPSIRPPVPAGLRGSSAWDQDPSRPRHPRAKDRIKARTNILGMVADTESSDSLPGSSNAASEMPANGSIHRKSQEKPPKKTHKAEREKLKRDQLNDLFVELGSMLDLDRQNTGKATVLGDAARVLRDLITQVESLRQEQSALVSERQYVSSEKNELQEENSSLKSQISELQTELCARMRSSSLSQTSIGMSDPATHQQMQMWSSIPHLSSVAMAARPASAASPLHGQEGYSADAGQAGYAPQPQPRELQLFPGSSASSSPERERSSRLGSGQATPPSLTDSLPGQLCLSLLQPSQEASGGGGGGVMSRSREERRDG* |
NA |
False |
49.5 |
52.8 |
NA |
NA |
57.2 |
52.3 |
45.9 |
50.1 |
|||
gene2 |
gene2_1 |
AGTGCTAGGTGTTGCTGCGCGAGTACTGCGAGATCTAATCACTCAAGTGGAATCTCTCAGGCAGGAGCAATCTGCTCTTG |
gene2 |
0 |
3751 |
3831 |
((3762, ‘A’, ‘T’), (3817, ‘A’, ‘G’)) |
() |
AGTGCTAGGTGATGCTGCGCGAGTACTGCGAGATCTAATCACTCAAGTGGAATCTCTCAGGCAGGAACAATCTGCTCTTG AGTGCTAGGTGTTGCTGCGCGAGTACTGCGAGATCTAATCACTCAAGTGGAATCTCTCAGGCAGGAGCAATCTGCTCTTG |
nan |
False |
False |
-31:S:A-T,24:S:A-G |
3795 |
False |
False |
False |
EFPCRWGRRRRHVSPRYRLGHTSVRVGGRHTLAAPACVCSPPTLPALALHFPWQSSLVPDFLPERIRPPSIRPPVPAGLRGSSAWDQDPSRPRHPRAKDRIKARTNILGMVADTESSDSLPGSSNAASEMPANGSIHRKSQEKPPKKTHKAEREKLKRDQLNDLFVELGSMLDLDRQNTGKATVLGDAARVLRDLITQVESLRQEQSALVSERQYVSSEKNELQEENSSLKSQISELQTELCARMRSSSLSQTSIGMSDPATHQQMQMWSSIPHLSSVAMAARPASAASPLHGQEGYSADAGQAGYAPQPQPRELQLFPGSSASSSPERERSSRLGSGQATPPSLTDSLPGQLCLSLLQPSQEASGGGGGGVMSRSREERRDG* |
100.0 |
False |
50.5 |
47.2 |
48.2 |
NA |
NA |
47.7 |
NA |
49.9 |
gene2 |
gene2_1 |
AGTGCTAGGTGATGCTGCGCGAGTACTGCGAGATCTAATCACAAGTGGAATCTCTCAGGCAGGAACAATCTGCTCTTG |
gene2 |
2 |
3751 |
3831 |
() |
((3792, ‘ACT’, ‘A’),) |
AGTGCTAGGTGATGCTGCGCGAGTACTGCGAGATCTAATCACTCAAGTGGAATCTCTCAGGCAGGAACAATCTGCTCTTG AGTGCTAGGTGATGCTGCGCGAGTACTGCGAGATCTAATCA–CAAGTGGAATCTCTCAGGCAGGAACAATCTGCTCTTG |
True |
nan |
True |
-1:2D:ACT-A |
3795 |
False |
False |
False |
EFPCRWGRRRRHVSPRYRLGHTSVRVGGRHTLAAPACVCSPPTLPALALHFPWQSSLVPDFLPERIRPPSIRPPVPAGLRGSSAWDQDPSRPRHPRAKDRIKARTNILGMVADTESSDSLPGSSNAASEMPANGSIHRKSQEKPPKKTHKAEREKLKRDQLNDLFVELGSMLDLDRQNTGKATVLGDAARVLRDLITSGISQAGTICSCIGAPICQFREE* |
54.8 |
True |
NA |
NA |
51.8 |
45.0 |
NA |
NA |
NA |
NA |
gene2 |
gene2_1 |
AGTGCTAGGTGTTGCTGCGCGAGTACTGCGAGATCTAATCAAAGTGGAATCTCTCAGGCAGGAGCAATCTGCTCTTG |
gene2 |
3 |
3751 |
3831 |
((3762, ‘A’, ‘T’), (3817, ‘A’, ‘G’)) |
((3792, ‘ACTC’, ‘A’),) |
AGTGCTAGGTGATGCTGCGCGAGTACTGCGAGATCTAATCACTCAAGTGGAATCTCTCAGGCAGGAACAATCTGCTCTTG AGTGCTAGGTGTTGCTGCGCGAGTACTGCGAGATCTAATCA—AAGTGGAATCTCTCAGGCAGGAGCAATCTGCTCTTG |
True |
False |
True |
-1:3D:ACTC-A,-31:S:A-T,24:S:A-G |
3795 |
False |
False |
False |
EFPCRWGRRRRHVSPRYRLGHTSVRVGGRHTLAAPACVCSPPTLPALALHFPWQSSLVPDFLPERIRPPSIRPPVPAGLRGSSAWDQDPSRPRHPRAKDRIKARTNILGMVADTESSDSLPGSSNAASEMPANGSIHRKSQEKPPKKTHKAEREKLKRDQLNDLFVELGSMLDLDRQNTGKATVLGDAARVLRDLIKVESLRQEQSALVSERQYVSSEKNELQEENSSLKSQISELQTELCARMRSSSLSQTSIGMSDPATHQQMQMWSSIPHLSSVAMAARPASAASPLHGQEGYSADAGQAGYAPQPQPRELQLFPGSSASSSPERERSSRLGSGQATPPSLTDSLPGQLCLSLLQPSQEASGGGGGGVMSRSREERRDG* |
99.5 |
False |
NA |
NA |
NA |
55.0 |
42.8 |
NA |
54.1 |
NA |
gene3 |
gene3_1 |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
ref |
3078 |
3157 |
nan |
nan |
nan |
nan |
nan |
nan |
ref |
3121 |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYGVKSLSDEHSQLLSGGGGMDASMDNSWRLLPSQTAATFQATSYPLFGALSGLDESTIASLPKTQREPLSFFGSDFVTPKQENQTLRPFFDEWPKSRDSWPELNEDNSLGSSATQLSISIPMAPSDFNTSSRSPNGIPSR* |
NA |
False |
100.0 |
97.0 |
44.3 |
NA |
48.0 |
NA |
4.2 |
5.0 |
|||
gene3 |
gene3_1 |
CTGACGTGCTCACGTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
0 |
3078 |
3157 |
((3091, ‘T’, ‘G’),) |
() |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTGCTCACGTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
nan |
False |
False |
28:S:T-G |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYGVKSLSDEHSQLLSGGGGMDASMDNSWRLLPSQTAATFQATSYPLFGALSGLDESTIASLPKTQREPLSFFGSDFVTPKQENQTLRPFFDEWPKSRDSWPELNEDNSLGSSATQLSISIPMAPSDFNTSSRSPNGIPSR* |
100.0 |
False |
NA |
3.0 |
NA |
NA |
NA |
NA |
NA |
NA |
gene3 |
gene3_1 |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACATCCAGCTCT |
gene3 |
0 |
3078 |
3157 |
((3148, ‘G’, ‘T’),) |
() |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACATCCAGCTCT |
nan |
False |
False |
-29:S:G-T |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYGVKSLSDEHSQLLSGGGGMDASMDNSWRLLPSQTAATFQATSYPLFGALSGLDESTIASLPKTQREPLSFFGSDFVTPKQENQTLRPFFDEWPKSRDSWPELNEDNSLGSSATQLSISIPMAPSDFNTSSRSPNGIPSR* |
100.0 |
False |
NA |
NA |
5.6 |
NA |
NA |
NA |
NA |
NA |
gene3 |
gene3_1 |
CTGACGTACTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
0 |
3078 |
3157 |
((3085, ‘G’, ‘A’),) |
() |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTACTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
nan |
False |
False |
34:S:G-A |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYGVKSLSDEHSQLLSGGGGMDASMDNSWRLLPSQTAATFQATSYPLFGALSGLDESTIASLPKTQREPLSFFGSDFVTPKQENQTLRPFFDEWPKSRDSWPELNEDNSLGSSATQLSISIPMAPSDFNTSSRSPNGIPSR* |
100.0 |
False |
NA |
NA |
NA |
NA |
4.4 |
NA |
NA |
NA |
gene3 |
gene3_1 |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
-1 |
3078 |
3157 |
() |
((3120, ‘C’, ‘CG’),) |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTAC-GGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
True |
nan |
True |
-1:1I:C-CG |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYGGEVSVGRAQPALVRRRRHGRVNGQLVAPVAVPNRRHVPGHKLPSVRRAERSGREHHRLAAQDAEGAPLLLRERLRDPEAGEPDAAPLLRRVAQVEGLVAGAERGQQPRLLGHPALHLHPHGALRLQHQLQIAEWNTVKVKPSIRRHTCFFFLLFFRFEPFVLRTFF* |
62.6 |
True |
NA |
NA |
47.9 |
55.5 |
47.6 |
40.3 |
47.5 |
56.0 |
gene3 |
gene3_1 |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGTGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
-1 |
3078 |
3157 |
() |
((3121, ‘G’, ‘GT’),) |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACG-GGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGTGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
True |
nan |
True |
-2:1I:G-GT |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYVGEVSVGRAQPALVRRRRHGRVNGQLVAPVAVPNRRHVPGHKLPSVRRAERSGREHHRLAAQDAEGAPLLLRERLRDPEAGEPDAAPLLRRVAQVEGLVAGAERGQQPRLLGHPALHLHPHGALRLQHQLQIAEWNTVKVKPSIRRHTCFFFLLFFRFEPFVLRTFF* |
63.1 |
True |
NA |
NA |
NA |
44.5 |
NA |
NA |
7.6 |
9.0 |
gene3 |
gene3_1 |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACTGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
0 |
3078 |
3157 |
((3120, ‘G’, ‘T’),) |
() |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACTGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
nan |
True |
True |
-1:S:G-T |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYWVKSLSDEHSQLLSGGGGMDASMDNSWRLLPSQTAATFQATSYPLFGALSGLDESTIASLPKTQREPLSFFGSDFVTPKQENQTLRPFFDEWPKSRDSWPELNEDNSLGSSATQLSISIPMAPSDFNTSSRSPNGIPSR* |
99.7 |
False |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
3.0 |
gene3 |
gene3_1 |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
1 |
3078 |
3157 |
() |
((3120, ‘CG’, ‘C’),) |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTAC-GGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
True |
nan |
True |
-1:1D:CG-C |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYG* |
63.8 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
24.6 |
22.0 |
gene3 |
gene3_1 |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGTGAAGTCTCTGTCGGACGAGCCCAGCCAGCTCT |
gene3 |
1 |
3078 |
3157 |
((3145, ‘A’, ‘C’),) |
((3120, ‘CG’, ‘C’),) |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTAC-GGGTGAAGTCTCTGTCGGACGAGCCCAGCCAGCTCT |
True |
False |
True |
-1:1D:CG-C,-26:S:A-C |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYG* |
63.8 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
3.0 |
gene3 |
gene3_1 |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
2 |
3078 |
3157 |
() |
((3120, ‘CGG’, ‘C’),) |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTAC–GGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
True |
nan |
True |
-1:2D:CGG-C |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYGEVSVGRAQPALVRRRRHGRVNGQLVAPVAVPNRRHVPGHKLPSVRRAERSGREHHRLAAQDAEGAPLLLRERLRDPEAGEPDAAPLLRRVAQVEGLVAGAERGQQPRLLGHPALHLHPHGALRLQHQLQIAEWNTVKVKPSIRRHTCFFFLLFFRFEPFVLRTFF* |
62.5 |
True |
NA |
NA |
2.2 |
NA |
NA |
59.7 |
4.2 |
2.0 |
gene3 |
gene3_1 |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
3 |
3078 |
3157 |
() |
((3120, ‘CGGG’, ‘C’),) |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTAC—GTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
True |
nan |
True |
-1:3D:CGGG-C |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYVKSLSDEHSQLLSGGGGMDASMDNSWRLLPSQTAATFQATSYPLFGALSGLDESTIASLPKTQREPLSFFGSDFVTPKQENQTLRPFFDEWPKSRDSWPELNEDNSLGSSATQLSISIPMAPSDFNTSSRSPNGIPSR* |
99.7 |
False |
NA |
NA |
NA |
NA |
NA |
NA |
4.2 |
NA |
gene3 |
gene3_1 |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
5 |
3078 |
3157 |
() |
((3120, ‘CGGGGT’, ‘C’),) |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTAC—–GAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
True |
nan |
True |
-1:5D:CGGGGT-C |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYEVSVGRAQPALVRRRRHGRVNGQLVAPVAVPNRRHVPGHKLPSVRRAERSGREHHRLAAQDAEGAPLLLRERLRDPEAGEPDAAPLLRRVAQVEGLVAGAERGQQPRLLGHPALHLHPHGALRLQHQLQIAEWNTVKVKPSIRRHTCFFFLLFFRFEPFVLRTFF* |
62.6 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
1.7 |
NA |
gene3 |
gene3_1 |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
8 |
3078 |
3157 |
() |
((3118, ‘TACGGGGTG’, ‘T’),) |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCT——–AAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
True |
nan |
True |
1:8D:TACGGGGTG-T |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSA* |
63.3 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
2.5 |
NA |
gene3 |
gene3_1 |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
10 |
3078 |
3157 |
() |
((3121, ‘GGGGTGAAGTC’, ‘G’),) |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACG———-TCTGTCGGACGAGCACAGCCAGCTCT |
True |
nan |
True |
-2:10D:GGGGTGAAGTC-G |
3121 |
False |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYVCRTSTASSCPAAAAWTRQWTTRGACCRPKPPPRSRPQATLCSAR* |
69.3 |
True |
NA |
NA |
NA |
NA |
47.6 |
NA |
NA |
NA |
gene3 |
gene3_1 |
CTGACGTTGCTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
gene3 |
36 |
3078 |
3157 |
() |
((3085, ‘TGCTCACTTGCTGACATACCTAGGTACTC’, ‘T’), (3116, ‘CCTACGGGG’, ‘C’)) |
CTGACGTGCTCACTTGCTGACATACCTAGGTACTCTGCCTACGGGGTGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT CTGACGT—————————-TGC——–TGAAGTCTCTGTCGGACGAGCACAGCCAGCTCT |
True |
nan |
True |
34:28D:TGCTCACTTGCTGACATACCTAGGTACTC-T,3:8D:CCTACGGGG-C |
3121 |
False |
True |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLX* |
62.2 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
3.5 |
NA |
gene4 |
gene4_1 |
GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGTGCTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGA |
gene4 |
ref |
827 |
905 |
nan |
nan |
nan |
nan |
nan |
nan |
ref |
861 |
MEGGQDVFLGAAARAPPPPPSCPFHGSATATRSGGAQMLSFSSNGVAGLGLCSGASKMQGVLSRVRRPFTPTQWMELEHQALIYKHFAVNAPVPSSLLLPIKRSLNPWSSLGSSSLGWAPFRSGSADAEPGRCRRTDGKKWRCSRDAVGDQKYCERHIKRGCHRSRKHVEGRKATPTTADPTMAVSGGSLLHSHAVAWQQQGKSSAANVTDPFSLGSNRNLLDKQNLGDQFSISTSMDSFDFSSSHSSPNQAKVAFSPVAMQHEHDQLYLVHGAGSSAENVNKSQDGQLLVSRETIDDGPLGEVFKGKSCQSASADILTDHWTSTRDLRPPTGILQMSSSNTVPAENHTSNSSYLMARMANSQTVPTLH* |
NA |
False |
100.0 |
98.8 |
48.3 |
64.8 |
98.4 |
98.7 |
53.6 |
41.9 |
|||
gene4 |
gene4_1 |
GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGTGCTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGAGAACCT |
gene4 |
-6 |
827 |
905 |
() |
((78, ‘A——’, ‘AGAACCT’),) |
GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGTGCTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGA—— GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGTGCTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGAGAACCT |
False |
nan |
False |
-781:0I:A——-AGAACCT |
861 |
False |
False |
False |
MEGGQDVFLGAAARAPPPPPSCPFHGSATATRSGGAQMLSFSSNGVAGLGLCSGASKMQGVLSRVRRPFTPTQWMELEHQALIYKHFAVNAPVPSSLLLPIKRSLNPWSSLGSSSLGWAPFRSGSADAEPGRCRRTDGKKWRCSRDAVGDQKYCERHIKRGCHRSRKHVEGRKATPTTADPTMAVSGGSLLHSHAVAWQQQGKSSAANVTDPFSLGSNRNLLDKQNLGDQFSISTSMDSFDFSSSHSSPNQAKVAFSPVAMQHEHDQLYLVHGAGSSAENVNKSQDGQLLVSRETIDDGPLGEVFKGKSCQSASADILTDHWTSTRDLRPPTGILQMSSSNTVPAENHTSNSSYLMARMANSQTVPTLH* |
100.0 |
False |
NA |
1.2 |
NA |
NA |
1.6 |
1.3 |
NA |
NA |
gene4 |
gene4_1 |
GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGTGACTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGA |
gene4 |
-1 |
827 |
905 |
() |
((861, ‘G’, ‘GA’),) |
GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGTG-CTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGA GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGTGACTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGA |
True |
nan |
True |
2:1I:G-GA |
861 |
False |
False |
False |
MEGGQDVFLGAAARAPPPPPSCPFHGSATATRSGGAQMLSFSSNGVAGLGL* |
13.8 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
1.5 |
gene4 |
gene4_1 |
GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGTGTCTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGA |
gene4 |
-1 |
827 |
905 |
() |
((861, ‘G’, ‘GT’),) |
GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGTG-CTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGA GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGTGTCTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGA |
True |
nan |
True |
2:1I:G-GT |
861 |
False |
False |
False |
MEGGQDVFLGAAARAPPPPPSCPFHGSATATRSGGAQMLSFSSNGVAGLGLCLRCQQDAGCVVEGEEALHSDAVDGAGAPGPDLQALRCECPCAVQLAPPYQKKPQSMEQPWLQLIGMGTISFRLC* |
23.6 |
True |
NA |
NA |
51.7 |
35.2 |
NA |
NA |
46.4 |
55.3 |
gene4 |
gene4_1 |
GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGTCTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGA |
gene4 |
1 |
827 |
905 |
() |
((860, ‘TG’, ‘T’),) |
GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGTGCTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGA GATGGTGTTCTTGTGATGAAGGGTTGGGTCTGT-CTCAGGTGCCAGCAAGATGCAGGGTGTGTTGTCGAGGGTGAGGA |
True |
nan |
True |
1:1D:TG-T |
861 |
False |
False |
False |
MEGGQDVFLGAAARAPPPPPSCPFHGSATATRSGGAQMLSFSSNGVAGLGLSQVPARCRVCCRG* |
16.5 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
1.3 |
gene5 |
gene5_1 |
CGCGCTGCCCATCCCTCCCCACCAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
gene5 |
ref |
1190 |
1272 |
nan |
nan |
nan |
nan |
nan |
nan |
ref |
1212 |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHQSLLLDEISSAANPSLQLLEQHGLGGMIPSGRNAAALHPHPPQPQPPAAGKGAKEPSPRANSAINRPPEKRQRVPSAYNRFIKDEIQRIKAGNPDISHREAFSAAAKNWAHFPHIHFGLMPDHQGPKKTSLLPQDHQRSDGGGLLKEGLYAAAANMGVAPY* |
NA |
False |
100.0 |
100.0 |
38.3 |
45.4 |
43.7 |
42.8 |
4.5 |
2.2 |
|||
gene5 |
gene5_1 |
CGCGCTGCCCATCCCTCCCCACACAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
gene5 |
-1 |
1190 |
1272 |
() |
((1212, ‘C’, ‘CA’),) |
CGCGCTGCCCATCCCTCCCCAC-CAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA CGCGCTGCCCATCCCTCCCCACACAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
True |
nan |
True |
-2:1I:C-CA |
1212 |
False |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHTIPPAGRDIQRREPEPAVAGAARPRRHDPQRQERGRAAPAPAPAPAARSG* |
57.4 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
1.7 |
gene5 |
gene5_1 |
CGCGCTGCCCATCCCTCCCCACTCAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
gene5 |
-1 |
1190 |
1272 |
() |
((1212, ‘C’, ‘CT’),) |
CGCGCTGCCCATCCCTCCCCAC-CAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA CGCGCTGCCCATCCCTCCCCACTCAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
True |
nan |
True |
-2:1I:C-CT |
1212 |
False |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHSIPPAGRDIQRREPEPAVAGAARPRRHDPQRQERGRAAPAPAPAPAARSG* |
57.4 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
1.2 |
3.0 |
gene5 |
gene5_1 |
CGCGCTGCCCATCCCTCCCCACAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
gene5 |
1 |
1190 |
1272 |
() |
((1211, ‘AC’, ‘A’),) |
CGCGCTGCCCATCCCTCCCCACCAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA CGCGCTGCCCATCCCTCCCCA-CAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
True |
nan |
True |
-1:1D:AC-A |
1212 |
False |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHNPSCWTRYPAPRTRACSCWSSTASAA* |
53.9 |
True |
NA |
NA |
2.1 |
NA |
NA |
NA |
12.3 |
18.5 |
gene5 |
gene5_1 |
CGCGCTGCCCATCCCTCCCCACATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
gene5 |
2 |
1190 |
1272 |
() |
((1212, ‘CCA’, ‘C’),) |
CGCGCTGCCCATCCCTCCCCACCAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA CGCGCTGCCCATCCCTCCCCAC–ATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
True |
nan |
True |
-2:2D:CCA-C |
1212 |
False |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHIPPAGRDIQRREPEPAVAGAARPRRHDPQRQERGRAAPAPAPAPAARSG* |
57.4 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
6.0 |
4.3 |
gene5 |
gene5_1 |
CGCGCTGCCCATCCCTCCCCAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
gene5 |
3 |
1190 |
1272 |
() |
((1208, ‘CCCA’, ‘C’),) |
CGCGCTGCCCATCCCTCCCCACCAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA CGCGCTGCCCATCCCTCC—CCAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
True |
nan |
True |
2:3D:CCCA-C |
1212 |
False |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPQSLLLDEISSAANPSLQLLEQHGLGGMIPSGRNAAALHPHPPQPQPPAAGKGAKEPSPRANSAINRPPEKRQRVPSAYNRFIKDEIQRIKAGNPDISHREAFSAAAKNWAHFPHIHFGLMPDHQGPKKTSLLPQDHQRSDGGGLLKEGLYAAAANMGVAPY* |
99.7 |
False |
NA |
NA |
NA |
NA |
NA |
NA |
1.7 |
45.5 |
gene5 |
gene5_1 |
CGCGCTGCCCATCCCTCCCCACTCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
gene5 |
3 |
1190 |
1272 |
() |
((1212, ‘CCAA’, ‘C’),) |
CGCGCTGCCCATCCCTCCCCACCAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA CGCGCTGCCCATCCCTCCCCAC—TCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
True |
nan |
True |
-2:3D:CCAA-C |
1212 |
False |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHSLLLDEISSAANPSLQLLEQHGLGGMIPSGRNAAALHPHPPQPQPPAAGKGAKEPSPRANSAINRPPEKRQRVPSAYNRFIKDEIQRIKAGNPDISHREAFSAAAKNWAHFPHIHFGLMPDHQGPKKTSLLPQDHQRSDGGGLLKEGLYAAAANMGVAPY* |
99.7 |
False |
NA |
NA |
NA |
54.6 |
NA |
NA |
1.9 |
2.3 |
gene5 |
gene5_1 |
CGCGCTGCCCATCCCTCCCCACCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
gene5 |
4 |
1190 |
1272 |
() |
((1212, ‘CCAAT’, ‘C’),) |
CGCGCTGCCCATCCCTCCCCACCAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA CGCGCTGCCCATCCCTCCCCAC—-CCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
True |
nan |
True |
-2:4D:CCAAT-C |
1212 |
False |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHPSCWTRYPAPRTRACSCWSSTASAA* |
53.6 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
21.3 |
gene5 |
gene5_1 |
CGCGCTGCCCATCCCTCCCCACCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
gene5 |
5 |
1190 |
1272 |
() |
((1211, ‘ACCAAT’, ‘A’),) |
CGCGCTGCCCATCCCTCCCCACCAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA CGCGCTGCCCATCCCTCCCCA—–CCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
True |
nan |
True |
-1:5D:ACCAAT-A |
1212 |
False |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHPPAGRDIQRREPEPAVAGAARPRRHDPQRQERGRAAPAPAPAPAARSG* |
57.1 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
1.2 |
1.2 |
gene5 |
gene5_1 |
CGCGCTGCCCATCCCTCCCCACGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
gene5 |
21 |
1190 |
1272 |
() |
((1211, ‘ACCAATCCCTCCTGCTGGTAAG’, ‘A’),) |
CGCGCTGCCCATCCCTCCCCACCAATCCCTCCTGCTGGTAAGCGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA CGCGCTGCCCATCCCTCCCCA———————CGCGCGGCCGGCGGAGAGCCGGCTGGGACTGGCACTGGGA |
True |
nan |
True |
-1:21D:ACCAATCCCTCCTGCTGGTAAG-A |
1212 |
False |
True |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPX* |
49.2 |
True |
NA |
NA |
59.6 |
NA |
56.3 |
57.2 |
71.2 |
NA |
gene6 |
gene6_1 |
GATGGAGGGAGACGACTTGTCATCCTTGTCGCCATCACCGGTCCCGTACATTTTCGGTGGTGGGTTGAGGGGAAGGA |
gene6 |
ref |
704 |
781 |
nan |
nan |
nan |
nan |
nan |
nan |
ref |
741 |
MDFPGGSGRRPQQQEPEHLPPMTPLPLARQGSVYSLTFDEFQSSLGGAAKDFGSMNMDELLRSIWSAEEVHSVAAASASAADHAHAAARGPVSIQHQGSLTLPRTLSQKTVDEVWRDLTCVGGGPSSGSAAPAAPPPPAQRHPTLGEITLEEFLVRAGVVREDMTAPPPVPPAPVCPAPAPRPPVLFPHGNVLAPLVPPLQFGNGFVSGAVGQQRGGPVPPAVSPRPVTASAFGKMEGDDLSSLSPSPVPYIFGGGLRGRKPPAMEKVVERRQRRMIKNRESAARSRQRKQKNPHGTGARLNGDGVAVTSVFGLDGEDHREGDQRRPKEAAEAHERSREARDVTRQKTGEYTDASREAAQEARDRSRATAQEGRHRRQGEGGQGRGRGHSSATRIWR* |
NA |
False |
100.0 |
96.7 |
52.6 |
NA |
100.0 |
50.2 |
100.0 |
100.0 |
|||
gene6 |
gene6_1 |
GATGGAGGGAGACGACTTGTCATCCTTGTCGCCATCACCGGCCCCGTACATTTTCGGTGGTGGGTTGAGGGGAAGGA |
gene6 |
0 |
704 |
781 |
((745, ‘T’, ‘C’),) |
() |
GATGGAGGGAGACGACTTGTCATCCTTGTCGCCATCACCGGTCCCGTACATTTTCGGTGGTGGGTTGAGGGGAAGGA GATGGAGGGAGACGACTTGTCATCCTTGTCGCCATCACCGGCCCCGTACATTTTCGGTGGTGGGTTGAGGGGAAGGA |
nan |
True |
True |
-6:S:T-C |
741 |
False |
False |
False |
MDFPGGSGRRPQQQEPEHLPPMTPLPLARQGSVYSLTFDEFQSSLGGAAKDFGSMNMDELLRSIWSAEEVHSVAAASASAADHAHAAARGPVSIQHQGSLTLPRTLSQKTVDEVWRDLTCVGGGPSSGSAAPAAPPPPAQRHPTLGEITLEEFLVRAGVVREDMTAPPPVPPAPVCPAPAPRPPVLFPHGNVLAPLVPPLQFGNGFVSGAVGQQRGGPVPPAVSPRPVTASAFGKMEGDDLSSLSPSPAPYIFGGGLRGRKPPAMEKVVERRQRRMIKNRESAARSRQRKQKNPHGTGARLNGDGVAVTSVFGLDGEDHREGDQRRPKEAAEAHERSREARDVTRQKTGEYTDASREAAQEARDRSRATAQEGRHRRQGEGGQGRGRGHSSATRIWR* |
99.7 |
False |
NA |
1.1 |
NA |
NA |
NA |
NA |
NA |
NA |
gene6 |
gene6_1 |
GATGGAGGGAGACGACTTGTCATCCTTGTCGCCATCACCGGTCCCATACATTTTCGGTGGTGGGTTGAGGGGAAGGA |
gene6 |
0 |
704 |
781 |
((749, ‘G’, ‘A’),) |
() |
GATGGAGGGAGACGACTTGTCATCCTTGTCGCCATCACCGGTCCCGTACATTTTCGGTGGTGGGTTGAGGGGAAGGA GATGGAGGGAGACGACTTGTCATCCTTGTCGCCATCACCGGTCCCATACATTTTCGGTGGTGGGTTGAGGGGAAGGA |
nan |
True |
True |
-10:S:G-A |
741 |
False |
False |
False |
MDFPGGSGRRPQQQEPEHLPPMTPLPLARQGSVYSLTFDEFQSSLGGAAKDFGSMNMDELLRSIWSAEEVHSVAAASASAADHAHAAARGPVSIQHQGSLTLPRTLSQKTVDEVWRDLTCVGGGPSSGSAAPAAPPPPAQRHPTLGEITLEEFLVRAGVVREDMTAPPPVPPAPVCPAPAPRPPVLFPHGNVLAPLVPPLQFGNGFVSGAVGQQRGGPVPPAVSPRPVTASAFGKMEGDDLSSLSPSPVPYIFGGGLRGRKPPAMEKVVERRQRRMIKNRESAARSRQRKQKNPHGTGARLNGDGVAVTSVFGLDGEDHREGDQRRPKEAAEAHERSREARDVTRQKTGEYTDASREAAQEARDRSRATAQEGRHRRQGEGGQGRGRGHSSATRIWR* |
100.0 |
False |
NA |
1.1 |
NA |
NA |
NA |
NA |
NA |
NA |
gene6 |
gene6_1 |
GATGGAGGGAGACGACTTGTCATCCTTGTCGCCATCACCGGTCCCGTACATTTTCTGTGGTGGGTTGAGGGGAAGGA |
gene6 |
0 |
704 |
781 |
((759, ‘G’, ‘T’),) |
() |
GATGGAGGGAGACGACTTGTCATCCTTGTCGCCATCACCGGTCCCGTACATTTTCGGTGGTGGGTTGAGGGGAAGGA GATGGAGGGAGACGACTTGTCATCCTTGTCGCCATCACCGGTCCCGTACATTTTCTGTGGTGGGTTGAGGGGAAGGA |
nan |
False |
False |
-20:S:G-T |
741 |
False |
False |
False |
MDFPGGSGRRPQQQEPEHLPPMTPLPLARQGSVYSLTFDEFQSSLGGAAKDFGSMNMDELLRSIWSAEEVHSVAAASASAADHAHAAARGPVSIQHQGSLTLPRTLSQKTVDEVWRDLTCVGGGPSSGSAAPAAPPPPAQRHPTLGEITLEEFLVRAGVVREDMTAPPPVPPAPVCPAPAPRPPVLFPHGNVLAPLVPPLQFGNGFVSGAVGQQRGGPVPPAVSPRPVTASAFGKMEGDDLSSLSPSPVPYIFGGGLRGRKPPAMEKVVERRQRRMIKNRESAARSRQRKQKNPHGTGARLNGDGVAVTSVFGLDGEDHREGDQRRPKEAAEAHERSREARDVTRQKTGEYTDASREAAQEARDRSRATAQEGRHRRQGEGGQGRGRGHSSATRIWR* |
100.0 |
False |
NA |
1.1 |
NA |
3.4 |
NA |
NA |
NA |
NA |
gene6 |
gene6_1 |
GATGGAGGGAGACGACTTGTCATCCTTGTCGCCATCCGGTCCCGTACATTTTCGGTGGTGGGTTGAGGGGAAGGA |
gene6 |
2 |
704 |
781 |
() |
((739, ‘TCA’, ‘T’),) |
GATGGAGGGAGACGACTTGTCATCCTTGTCGCCATCACCGGTCCCGTACATTTTCGGTGGTGGGTTGAGGGGAAGGA GATGGAGGGAGACGACTTGTCATCCTTGTCGCCAT–CCGGTCCCGTACATTTTCGGTGGTGGGTTGAGGGGAAGGA |
True |
nan |
True |
0:2D:TCA-T |
741 |
False |
False |
False |
MDFPGGSGRRPQQQEPEHLPPMTPLPLARQGSVYSLTFDEFQSSLGGAAKDFGSMNMDELLRSIWSAEEVHSVAAASASAADHAHAAARGPVSIQHQGSLTLPRTLSQKTVDEVWRDLTCVGGGPSSGSAAPAAPPPPAQRHPTLGEITLEEFLVRAGVVREDMTAPPPVPPAPVCPAPAPRPPVLFPHGNVLAPLVPPLQFGNGFVSGAVGQQRGGPVPPAVSPRPVTASAFGKMEGDDLSSLSPSGPVHFRWWVEGKEATGYGEGG* |
65.8 |
True |
NA |
NA |
47.4 |
96.6 |
NA |
49.8 |
NA |
NA |
gene7 |
gene7_1 |
TTTGCTGACCGACGTCACGTGCTGCAGGGCGCGGGCCGTGGGCTGGCCGCCGGTCCGCGCGTACCGGCGCAACGCGCTGCGCGACG |
gene7 |
ref |
304 |
390 |
nan |
nan |
nan |
nan |
nan |
nan |
ref |
339 |
MQQDLRNSERRNPEQAHPVMSASSTNSAASPAVSGLDYDDTALTLALPGSSAEPAADRKRAHADHDKPPSPKARAVGWPPVRAYRRNALRDEARLVKVAVDGAPYLRKVDLAAHDGYAALLRALHGMFASCLVAGAGADGAGRIDTAAEYMPTYEDKDGDWMLVGDVPFKMFVDSCKRIRLMKSSEAVNLSPRTSSRQ* |
NA |
False |
100.0 |
100.0 |
41.8 |
53.1 |
49.2 |
51.1 |
44.0 |
44.4 |
|||
gene7 |
gene7_1 |
TTTGCTGACCGACGTCACGTGCTGCAGGGCGCGGGACCGTGGGCTGGCCGCCGGTCCGCGCGTACCGGCGCAACGCGCTGCGCGACG |
gene7 |
-1 |
304 |
390 |
() |
((339, ‘G’, ‘GA’),) |
TTTGCTGACCGACGTCACGTGCTGCAGGGCGCGGG-CCGTGGGCTGGCCGCCGGTCCGCGCGTACCGGCGCAACGCGCTGCGCGACG TTTGCTGACCGACGTCACGTGCTGCAGGGCGCGGGACCGTGGGCTGGCCGCCGGTCCGCGCGTACCGGCGCAACGCGCTGCGCGACG |
True |
nan |
True |
2:1I:G-GA |
339 |
False |
False |
False |
MQQDLRNSERRNPEQAHPVMSASSTNSAASPAVSGLDYDDTALTLALPGSSAEPAADRKRAHADHDKPPSPKARDRGLAAGPRVPAQRAARRGQAREGGRGRRAVPAEGGPRGARRVRGPAPRAPRHVRLLPRCRSRSRRGGADRHRRRVHAHLRGQGRRLDARRRRPLQDVRGLVQEDPPHEELRGRQPISEDIIPAVIVVGVDAICPTRLISPN* |
49.0 |
True |
NA |
NA |
58.2 |
46.9 |
50.8 |
48.9 |
1.4 |
55.6 |
gene7 |
gene7_1 |
TTTGCTGACCGACGTCACGTGCTGCAGGGCGCGGCCGTGGGCTGGCCGCCGGTCCGCGCGTACCGGCGCAACGCGCTGCGCGACG |
gene7 |
1 |
304 |
390 |
() |
((336, ‘CG’, ‘C’),) |
TTTGCTGACCGACGTCACGTGCTGCAGGGCGCGGGCCGTGGGCTGGCCGCCGGTCCGCGCGTACCGGCGCAACGCGCTGCGCGACG TTTGCTGACCGACGTCACGTGCTGCAGGGCGC-GGCCGTGGGCTGGCCGCCGGTCCGCGCGTACCGGCGCAACGCGCTGCGCGACG |
True |
nan |
True |
-1:1D:CG-C |
339 |
False |
False |
False |
MQQDLRNSERRNPEQAHPVMSASSTNSAASPAVSGLDYDDTALTLALPGSSAEPAADRKRAHADHDKPPSPKARPWAGRRSARTGATRCATRPGS* |
43.4 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
54.6 |
NA |
gene8 |
gene8_1 |
GCTGAATATTTTTTTCTCTTTGGTTTGTTGCTGTTGTTGTTGTTGGCTGATGCAGGGCTTCAGGAAGATAGTGGCGGACAGGTGGGA |
gene8 |
ref |
1767 |
1854 |
nan |
nan |
nan |
nan |
nan |
nan |
ref |
1825 |
MAFLVERCGEMVVSMESPHAKPVPAPFLTKTYQLVDDPCTDHIVSWGDDDTTFVVWRPPEFARDLLPNYFKHNNFSSFVRQLNTYGFRKIVADRWEFANEFFRKGAKHLLAEIHRRKSSQPLPTPMPPHQPYHHHLHHLHHHLSPFSPPPLAQPVPSYHHHHFQEEPIATATAPHGGAQAGAAGGGNNEGSGAGSRRGLSGRANHVAPVTSPSSAAHASLPSAAGGGAAASSCRLMELDPADSPSPPRRPEADDGTDTVKLFGVALQGKKKKRAHQEDGDDGNHEQGSSDV* |
NA |
False |
100.0 |
98.4 |
55.0 |
65.5 |
54.6 |
52.5 |
63.6 |
51.5 |
|||
gene8 |
gene8_1 |
GCTGAATATTTTTTTCTCTTTGGTTTGTTGCTGTTGTTGTTGTTGGCTGATGCAGGGCTTTCAGGAAGATAGTGGCGGACAGGTGGGA |
gene8 |
-1 |
1767 |
1854 |
() |
((1825, ‘C’, ‘CT’),) |
GCTGAATATTTTTTTCTCTTTGGTTTGTTGCTGTTGTTGTTGTTGGCTGATGCAGGGC-TTCAGGAAGATAGTGGCGGACAGGTGGGA GCTGAATATTTTTTTCTCTTTGGTTTGTTGCTGTTGTTGTTGTTGGCTGATGCAGGGCTTTCAGGAAGATAGTGGCGGACAGGTGGGA |
True |
nan |
True |
2:1I:C-CT |
1825 |
False |
False |
False |
MAFLVERCGEMVVSMESPHAKPVPAPFLTKTYQLVDDPCTDHIVSWGDDDTTFVVWRPPEFARDLLPNYFKHNNFSSFVRQLNTYGFQEDSGGQVGVRQRVLQEGRQAPTRRDPPEEVVAAAADADAAAPALPPPPPPSPPPPQPVLPAAAGTAGAVVPPPPLPRRAHRHRHRAARRCSSRRRRWRQQ* |
39.1 |
True |
NA |
1.6 |
45.0 |
34.5 |
45.4 |
47.5 |
36.4 |
48.5 |
gene9 |
gene9_1 |
GGCGGCAAGCGCCTCCGCCCCGTGCTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
gene9 |
ref |
315 |
402 |
nan |
nan |
nan |
nan |
nan |
nan |
ref |
339 |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVLAIAACELVGGTAAAAVPVACAVEMIHTASLIHDDMPCMDDDALRRGRPSNHVAFGEPTALLAGDALLALAFEHVARGSAGAGVPADRALRAVVELGSVAGVGGIAAGQVADMASEGAPSGSVSLAALEYIHVHKTARLVEAAAVSGAVVGGGGDGEVERVRRYAHFLGLLGQVVDDVLDVTGTSEQLGKTAGKDVAAGKATYPRLMGLKGARAYMGELLAKAEAELDGLDAAPTAPLRHLARGGDHIDVMVVVGYDWVGFGVGIG* |
NA |
False |
100.0 |
100.0 |
49.3 |
55.0 |
53.7 |
54.2 |
29.6 |
38.5 |
|||
gene9 |
gene9_1 |
GGCGGCAAGCGCCTCCGCCCCGTGACTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
gene9 |
-1 |
315 |
402 |
() |
((339, ‘G’, ‘GA’),) |
GGCGGCAAGCGCCTCCGCCCCGTG-CTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC GGCGGCAAGCGCCTCCGCCCCGTGACTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
True |
nan |
True |
-2:1I:G-GA |
339 |
False |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVTGHRRVRARGRDRGRGRPGGVRRRDDPHRVAHPRRHAVHGRRRAPPRPPLQPRRVRRAHGATRRRRAAGARFRARRPRQRGRRRPRGPRAPRRRGARERSWRRRHRRGAGRRHGERGSPLRLREPGRAGVHPCA* |
38.9 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
5.6 |
8.9 |
gene9 |
gene9_1 |
GGCGGCAAGCGCCTCCGCCCCGTGCCTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
gene9 |
-1 |
315 |
402 |
() |
((339, ‘G’, ‘GC’),) |
GGCGGCAAGCGCCTCCGCCCCGTG-CTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC GGCGGCAAGCGCCTCCGCCCCGTGCCTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
True |
nan |
True |
-2:1I:G-GC |
339 |
False |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVPGHRRVRARGRDRGRGRPGGVRRRDDPHRVAHPRRHAVHGRRRAPPRPPLQPRRVRRAHGATRRRRAAGARFRARRPRQRGRRRPRGPRAPRRRGARERSWRRRHRRGAGRRHGERGSPLRLREPGRAGVHPCA* |
38.9 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
1.2 |
3.0 |
gene9 |
gene9_1 |
GGCGGCAAGCGCCTCCGCCCCGTGTCTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
gene9 |
-1 |
315 |
402 |
() |
((339, ‘G’, ‘GT’),) |
GGCGGCAAGCGCCTCCGCCCCGTG-CTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC GGCGGCAAGCGCCTCCGCCCCGTGTCTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
True |
nan |
True |
-2:1I:G-GT |
339 |
False |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVSGHRRVRARGRDRGRGRPGGVRRRDDPHRVAHPRRHAVHGRRRAPPRPPLQPRRVRRAHGATRRRRAAGARFRARRPRQRGRRRPRGPRAPRRRGARERSWRRRHRRGAGRRHGERGSPLRLREPGRAGVHPCA* |
38.9 |
True |
NA |
NA |
50.7 |
45.0 |
NA |
NA |
56.5 |
1.1 |
gene9 |
gene9_1 |
GGCGGCAAGCGCCTCCGCCCCGTGTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
gene9 |
1 |
315 |
402 |
() |
((339, ‘GC’, ‘G’),) |
GGCGGCAAGCGCCTCCGCCCCGTGCTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC GGCGGCAAGCGCCTCCGCCCCGTG-TGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
True |
nan |
True |
-2:1D:GC-G |
339 |
False |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVWPSPRASSWAGPRPRPSRWRAPSR* |
34.0 |
True |
NA |
NA |
NA |
NA |
46.3 |
NA |
2.3 |
2.4 |
gene9 |
gene9_1 |
GGCGGCAAGCGCCTCCGCCCCGTGGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
gene9 |
2 |
315 |
402 |
() |
((339, ‘GCT’, ‘G’),) |
GGCGGCAAGCGCCTCCGCCCCGTGCTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC GGCGGCAAGCGCCTCCGCCCCGTG–GGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
True |
nan |
True |
-2:2D:GCT-G |
339 |
False |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVGHRRVRARGRDRGRGRPGGVRRRDDPHRVAHPRRHAVHGRRRAPPRPPLQPRRVRRAHGATRRRRAAGARFRARRPRQRGRRRPRGPRAPRRRGARERSWRRRHRRGAGRRHGERGSPLRLREPGRAGVHPCA* |
38.9 |
True |
NA |
NA |
NA |
NA |
NA |
45.8 |
2.5 |
1.9 |
gene9 |
gene9_1 |
GGCGGCAAGCGCCTCCGCCCCGTGTGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
gene9 |
2 |
315 |
402 |
() |
((339, ‘GC’, ‘G’), (341, ‘TG’, ‘T’)) |
GGCGGCAAGCGCCTCCGCCCCGTGCTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC GGCGGCAAGCGCCTCCGCCCCGTG-T-GCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
True |
nan |
True |
-2:1D:GC-G,-4:1D:TG-T |
339 |
False |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVCHRRVRARGRDRGRGRPGGVRRRDDPHRVAHPRRHAVHGRRRAPPRPPLQPRRVRRAHGATRRRRAAGARFRARRPRQRGRRRPRGPRAPRRRGARERSWRRRHRRGAGRRHGERGSPLRLREPGRAGVHPCA* |
38.9 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
42.1 |
gene9 |
gene9_1 |
GGCGGCAAGCGCCTCCGCCCCGTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
gene9 |
3 |
315 |
402 |
() |
((337, ‘GTGC’, ‘G’),) |
GGCGGCAAGCGCCTCCGCCCCGTGCTGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC GGCGGCAAGCGCCTCCGCCCCG—TGGCCATCGCCGCGTGCGAGCTCGTGGGCGGGACCGCGGCCGCGGCCGTCCCGGTGGCGTGC |
True |
nan |
True |
0:3D:GTGC-G |
339 |
False |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVAIAACELVGGTAAAAVPVACAVEMIHTASLIHDDMPCMDDDALRRGRPSNHVAFGEPTALLAGDALLALAFEHVARGSAGAGVPADRALRAVVELGSVAGVGGIAAGQVADMASEGAPSGSVSLAALEYIHVHKTARLVEAAAVSGAVVGGGGDGEVERVRRYAHFLGLLGQVVDDVLDVTGTSEQLGKTAGKDVAAGKATYPRLMGLKGARAYMGELLAKAEAELDGLDAAPTAPLRHLARGGDHIDVMVVVGYDWVGFGVGIG* |
99.7 |
False |
NA |
NA |
NA |
NA |
NA |
NA |
2.3 |
2.1 |
Haplotype |
Reference |
Locus |
Target |
Expected cut site |
FILTER_gRNA |
atgCheck |
splicingSiteCheck |
protein_sequence |
pairwiseProteinIdentity (%) |
Effect |
sample1 |
sample2 |
sample3 |
sample4 |
sample5 |
sample6 |
sample7 |
sample8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
gene1_1_ref |
gene1 |
gene1_1 |
gene1 |
1817 |
False |
MGSSYDPYPSPGADDLFLYLSDLGPASPSAYLDLPPTPQPQPYPQSQQQQQGSKGPTQDMLLPYISSMLMEDDIDDTFFYDYPDNPALLQAQQSFLDILSDDASSPTTTTGTTNSSASVNHSSSDASASAPPTPAAVDSYSPAPAVQFDGFDLDPAAFFSNGANSDLMSSAFLKGMEEANKFLPSQDKLVIDLDPPDDTKRFVLPTRAAENLAPGFNAAATTVPAAVAMAVKEEEVILAALDAALGSGGVVLGRGRRNRLDDDEEDLELQRRSTKQSALQGDGDERDVFEKYIMTCPETCTEQMQQLRIAMQEEAAKEAAVAAGNGKAKGRRGGREVVDLRTLLVHCAQAVASDDRRSATELLRQIKQHASPQGDATQRLAHCFAEGLQARLAGTGSMVYQSLMAKRTSAADILQAYQLYMAAICFKRVVFVFSNNTIYNAALGKMKIHIVDYGIHYGFQWPCFLRWIADREGGPPEVRITGIDLPQPGFRPTQRIEETGRRLSKYAQQFGVPFKYQAIAASKMESIRAEDLNLDPEEVLIVNCLYQFKNLMDESVVIESPRDIVLNNIRKMRPHTFIHAIVNGSFSAPFFVTRFREALFFYSALFDALDTTTPRDSNQRMLIEENLFGRAALNVIACEGTDRVERPETYKQWQVRNQRAGLKQQPLNPDVVQQDIGTSNQEEICDASMS* |
NA |
False |
100.0 |
100.0 |
100.0 |
100.0 |
100.0 |
100.0 |
100.0 |
100.0 |
||
gene2_1_ref |
gene2 |
gene2_1 |
gene2 |
3795 |
False |
EFPCRWGRRRRHVSPRYRLGHTSVRVGGRHTLAAPACVCSPPTLPALALHFPWQSSLVPDFLPERIRPPSIRPPVPAGLRGSSAWDQDPSRPRHPRAKDRIKARTNILGMVADTESSDSLPGSSNAASEMPANGSIHRKSQEKPPKKTHKAEREKLKRDQLNDLFVELGSMLDLDRQNTGKATVLGDAARVLRDLITQVESLRQEQSALVSERQYVSSEKNELQEENSSLKSQISELQTELCARMRSSSLSQTSIGMSDPATHQQMQMWSSIPHLSSVAMAARPASAASPLHGQEGYSADAGQAGYAPQPQPRELQLFPGSSASSSPERERSSRLGSGQATPPSLTDSLPGQLCLSLLQPSQEASGGGGGGVMSRSREERRDG* |
NA |
False |
100.0 |
100.0 |
48.2 |
NA |
57.2 |
100.0 |
45.9 |
100.0 |
||
gene2_1_-1:2D:ACT-A |
gene2 |
gene2_1 |
gene2 |
3795 |
True |
False |
False |
EFPCRWGRRRRHVSPRYRLGHTSVRVGGRHTLAAPACVCSPPTLPALALHFPWQSSLVPDFLPERIRPPSIRPPVPAGLRGSSAWDQDPSRPRHPRAKDRIKARTNILGMVADTESSDSLPGSSNAASEMPANGSIHRKSQEKPPKKTHKAEREKLKRDQLNDLFVELGSMLDLDRQNTGKATVLGDAARVLRDLITSGISQAGTICSCIGAPICQFREE* |
54.8 |
True |
NA |
NA |
51.8 |
45.0 |
NA |
NA |
NA |
NA |
gene2_1_-1:3D:ACTC-A |
gene2 |
gene2_1 |
gene2 |
3795 |
True |
False |
False |
EFPCRWGRRRRHVSPRYRLGHTSVRVGGRHTLAAPACVCSPPTLPALALHFPWQSSLVPDFLPERIRPPSIRPPVPAGLRGSSAWDQDPSRPRHPRAKDRIKARTNILGMVADTESSDSLPGSSNAASEMPANGSIHRKSQEKPPKKTHKAEREKLKRDQLNDLFVELGSMLDLDRQNTGKATVLGDAARVLRDLIKVESLRQEQSALVSERQYVSSEKNELQEENSSLKSQISELQTELCARMRSSSLSQTSIGMSDPATHQQMQMWSSIPHLSSVAMAARPASAASPLHGQEGYSADAGQAGYAPQPQPRELQLFPGSSASSSPERERSSRLGSGQATPPSLTDSLPGQLCLSLLQPSQEASGGGGGGVMSRSREERRDG* |
99.5 |
False |
NA |
NA |
NA |
55.0 |
42.8 |
NA |
54.1 |
NA |
gene3_1_ref |
gene3 |
gene3_1 |
gene3 |
3121 |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYGVKSLSDEHSQLLSGGGGMDASMDNSWRLLPSQTAATFQATSYPLFGALSGLDESTIASLPKTQREPLSFFGSDFVTPKQENQTLRPFFDEWPKSRDSWPELNEDNSLGSSATQLSISIPMAPSDFNTSSRSPNGIPSR* |
NA |
False |
100.0 |
100.0 |
49.9 |
NA |
52.4 |
NA |
4.2 |
5.0 |
||
gene3_1_-1:1D:CG-C |
gene3 |
gene3_1 |
gene3 |
3121 |
True |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYG* |
63.8 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
24.6 |
25.0 |
gene3_1_-1:1I:C-CG |
gene3 |
gene3_1 |
gene3 |
3121 |
True |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYGGEVSVGRAQPALVRRRRHGRVNGQLVAPVAVPNRRHVPGHKLPSVRRAERSGREHHRLAAQDAEGAPLLLRERLRDPEAGEPDAAPLLRRVAQVEGLVAGAERGQQPRLLGHPALHLHPHGALRLQHQLQIAEWNTVKVKPSIRRHTCFFFLLFFRFEPFVLRTFF* |
62.6 |
True |
NA |
NA |
47.9 |
55.5 |
47.6 |
40.3 |
47.5 |
56.0 |
gene3_1_-1:2D:CGG-C |
gene3 |
gene3_1 |
gene3 |
3121 |
True |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYGEVSVGRAQPALVRRRRHGRVNGQLVAPVAVPNRRHVPGHKLPSVRRAERSGREHHRLAAQDAEGAPLLLRERLRDPEAGEPDAAPLLRRVAQVEGLVAGAERGQQPRLLGHPALHLHPHGALRLQHQLQIAEWNTVKVKPSIRRHTCFFFLLFFRFEPFVLRTFF* |
62.5 |
True |
NA |
NA |
2.2 |
NA |
NA |
59.7 |
4.2 |
2.0 |
gene3_1_-1:3D:CGGG-C |
gene3 |
gene3_1 |
gene3 |
3121 |
True |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYVKSLSDEHSQLLSGGGGMDASMDNSWRLLPSQTAATFQATSYPLFGALSGLDESTIASLPKTQREPLSFFGSDFVTPKQENQTLRPFFDEWPKSRDSWPELNEDNSLGSSATQLSISIPMAPSDFNTSSRSPNGIPSR* |
99.7 |
False |
NA |
NA |
NA |
NA |
NA |
NA |
4.2 |
NA |
gene3_1_-1:5D:CGGGGT-C |
gene3 |
gene3_1 |
gene3 |
3121 |
True |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYEVSVGRAQPALVRRRRHGRVNGQLVAPVAVPNRRHVPGHKLPSVRRAERSGREHHRLAAQDAEGAPLLLRERLRDPEAGEPDAAPLLRRVAQVEGLVAGAERGQQPRLLGHPALHLHPHGALRLQHQLQIAEWNTVKVKPSIRRHTCFFFLLFFRFEPFVLRTFF* |
62.6 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
1.7 |
NA |
gene3_1_-1:S:G-T |
gene3 |
gene3_1 |
gene3 |
3121 |
True |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYWVKSLSDEHSQLLSGGGGMDASMDNSWRLLPSQTAATFQATSYPLFGALSGLDESTIASLPKTQREPLSFFGSDFVTPKQENQTLRPFFDEWPKSRDSWPELNEDNSLGSSATQLSISIPMAPSDFNTSSRSPNGIPSR* |
99.7 |
False |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
3.0 |
gene3_1_-2:10D:GGGGTGAAGTC-G |
gene3 |
gene3_1 |
gene3 |
3121 |
True |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYVCRTSTASSCPAAAAWTRQWTTRGACCRPKPPPRSRPQATLCSAR* |
69.3 |
True |
NA |
NA |
NA |
NA |
47.6 |
NA |
NA |
NA |
gene3_1_-2:1I:G-GT |
gene3 |
gene3_1 |
gene3 |
3121 |
True |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSAYVGEVSVGRAQPALVRRRRHGRVNGQLVAPVAVPNRRHVPGHKLPSVRRAERSGREHHRLAAQDAEGAPLLLRERLRDPEAGEPDAAPLLRRVAQVEGLVAGAERGQQPRLLGHPALHLHPHGALRLQHQLQIAEWNTVKVKPSIRRHTCFFFLLFFRFEPFVLRTFF* |
63.1 |
True |
NA |
NA |
NA |
44.5 |
NA |
NA |
7.6 |
9.0 |
gene3_1_1:8D:TACGGGGTG-T |
gene3 |
gene3_1 |
gene3 |
3121 |
True |
False |
False |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLRYSA* |
63.3 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
2.5 |
NA |
gene3_1_34:28D:TGCTCACTTGCTGACATACCTAGGTACTC-T,3:8D:CCTACGGGG-C |
gene3 |
gene3_1 |
gene3 |
3121 |
True |
False |
True |
MAMPFASLSPAADHRPSSLLPYCRAAPLSAVGEDAAAQAQQQQQHAMSGRWAARPPALFTAAQYEELEHQALIYKYLVAGVPVPPDLLLPLRRGFVYHQPALGYGPYFGKKVDPEPGRCRRTDGKKWRCSKEAAPDSKYCERHMHRGRNRSRKPVEAQLVPPPHAQQQQQQQAPAPTAGFQSHPMYPSILAGNGGGGGGVGGGAGGGGTFGLGPTSQLHMDSAAAYATAAGGGSKDLX* |
62.2 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
3.5 |
NA |
gene4_1_ref |
gene4 |
gene4_1 |
gene4 |
861 |
False |
MEGGQDVFLGAAARAPPPPPSCPFHGSATATRSGGAQMLSFSSNGVAGLGLCSGASKMQGVLSRVRRPFTPTQWMELEHQALIYKHFAVNAPVPSSLLLPIKRSLNPWSSLGSSSLGWAPFRSGSADAEPGRCRRTDGKKWRCSRDAVGDQKYCERHIKRGCHRSRKHVEGRKATPTTADPTMAVSGGSLLHSHAVAWQQQGKSSAANVTDPFSLGSNRNLLDKQNLGDQFSISTSMDSFDFSSSHSSPNQAKVAFSPVAMQHEHDQLYLVHGAGSSAENVNKSQDGQLLVSRETIDDGPLGEVFKGKSCQSASADILTDHWTSTRDLRPPTGILQMSSSNTVPAENHTSNSSYLMARMANSQTVPTLH* |
NA |
False |
100.0 |
100.0 |
48.3 |
64.8 |
100.0 |
100.0 |
53.6 |
41.9 |
||
gene4_1_1:1D:TG-T |
gene4 |
gene4_1 |
gene4 |
861 |
True |
False |
False |
MEGGQDVFLGAAARAPPPPPSCPFHGSATATRSGGAQMLSFSSNGVAGLGLSQVPARCRVCCRG* |
16.5 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
1.3 |
gene4_1_2:1I:G-GA |
gene4 |
gene4_1 |
gene4 |
861 |
True |
False |
False |
MEGGQDVFLGAAARAPPPPPSCPFHGSATATRSGGAQMLSFSSNGVAGLGL* |
13.8 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
1.5 |
gene4_1_2:1I:G-GT |
gene4 |
gene4_1 |
gene4 |
861 |
True |
False |
False |
MEGGQDVFLGAAARAPPPPPSCPFHGSATATRSGGAQMLSFSSNGVAGLGLCLRCQQDAGCVVEGEEALHSDAVDGAGAPGPDLQALRCECPCAVQLAPPYQKKPQSMEQPWLQLIGMGTISFRLC* |
23.6 |
True |
NA |
NA |
51.7 |
35.2 |
NA |
NA |
46.4 |
55.3 |
gene5_1_ref |
gene5 |
gene5_1 |
gene5 |
1212 |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHQSLLLDEISSAANPSLQLLEQHGLGGMIPSGRNAAALHPHPPQPQPPAAGKGAKEPSPRANSAINRPPEKRQRVPSAYNRFIKDEIQRIKAGNPDISHREAFSAAAKNWAHFPHIHFGLMPDHQGPKKTSLLPQDHQRSDGGGLLKEGLYAAAANMGVAPY* |
NA |
False |
100.0 |
100.0 |
38.3 |
45.4 |
43.7 |
42.8 |
4.5 |
2.2 |
||
gene5_1_-1:1D:AC-A |
gene5 |
gene5_1 |
gene5 |
1212 |
True |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHNPSCWTRYPAPRTRACSCWSSTASAA* |
53.9 |
True |
NA |
NA |
2.1 |
NA |
NA |
NA |
12.3 |
18.5 |
gene5_1_-1:21D:ACCAATCCCTCCTGCTGGTAAG-A |
gene5 |
gene5_1 |
gene5 |
1212 |
True |
False |
True |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPX* |
49.2 |
True |
NA |
NA |
59.6 |
NA |
56.3 |
57.2 |
71.2 |
NA |
gene5_1_-1:5D:ACCAAT-A |
gene5 |
gene5_1 |
gene5 |
1212 |
True |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHPPAGRDIQRREPEPAVAGAARPRRHDPQRQERGRAAPAPAPAPAARSG* |
57.1 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
1.2 |
1.2 |
gene5_1_-2:1I:C-CA |
gene5 |
gene5_1 |
gene5 |
1212 |
True |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHTIPPAGRDIQRREPEPAVAGAARPRRHDPQRQERGRAAPAPAPAPAARSG* |
57.4 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
1.7 |
gene5_1_-2:1I:C-CT |
gene5 |
gene5_1 |
gene5 |
1212 |
True |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHSIPPAGRDIQRREPEPAVAGAARPRRHDPQRQERGRAAPAPAPAPAARSG* |
57.4 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
1.2 |
3.0 |
gene5_1_-2:2D:CCA-C |
gene5 |
gene5_1 |
gene5 |
1212 |
True |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHIPPAGRDIQRREPEPAVAGAARPRRHDPQRQERGRAAPAPAPAPAARSG* |
57.4 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
6.0 |
4.3 |
gene5_1_-2:3D:CCAA-C |
gene5 |
gene5_1 |
gene5 |
1212 |
True |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHSLLLDEISSAANPSLQLLEQHGLGGMIPSGRNAAALHPHPPQPQPPAAGKGAKEPSPRANSAINRPPEKRQRVPSAYNRFIKDEIQRIKAGNPDISHREAFSAAAKNWAHFPHIHFGLMPDHQGPKKTSLLPQDHQRSDGGGLLKEGLYAAAANMGVAPY* |
99.7 |
False |
NA |
NA |
NA |
54.6 |
NA |
NA |
1.9 |
2.3 |
gene5_1_-2:4D:CCAAT-C |
gene5 |
gene5_1 |
gene5 |
1212 |
True |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPHPSCWTRYPAPRTRACSCWSSTASAA* |
53.6 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
21.3 |
gene5_1_2:3D:CCCA-C |
gene5 |
gene5_1 |
gene5 |
1212 |
True |
False |
False |
MSSSSSSSSAATVFPPSPQLPPPLLVENLPPLHQLTPPVAAAAAPASEQLCYVHCHFCDTVLVVSVPTSSLFKTVTVRCGHCSSLLTVNMRGLLFPGTPANTAAAAAAAPPPPPAAVTSTTATITTAPAPPPATSVNNNGQFHFIPHSLDLALPIPPQSLLLDEISSAANPSLQLLEQHGLGGMIPSGRNAAALHPHPPQPQPPAAGKGAKEPSPRANSAINRPPEKRQRVPSAYNRFIKDEIQRIKAGNPDISHREAFSAAAKNWAHFPHIHFGLMPDHQGPKKTSLLPQDHQRSDGGGLLKEGLYAAAANMGVAPY* |
99.7 |
False |
NA |
NA |
NA |
NA |
NA |
NA |
1.7 |
45.5 |
gene6_1_ref |
gene6 |
gene6_1 |
gene6 |
741 |
False |
MDFPGGSGRRPQQQEPEHLPPMTPLPLARQGSVYSLTFDEFQSSLGGAAKDFGSMNMDELLRSIWSAEEVHSVAAASASAADHAHAAARGPVSIQHQGSLTLPRTLSQKTVDEVWRDLTCVGGGPSSGSAAPAAPPPPAQRHPTLGEITLEEFLVRAGVVREDMTAPPPVPPAPVCPAPAPRPPVLFPHGNVLAPLVPPLQFGNGFVSGAVGQQRGGPVPPAVSPRPVTASAFGKMEGDDLSSLSPSPVPYIFGGGLRGRKPPAMEKVVERRQRRMIKNRESAARSRQRKQKNPHGTGARLNGDGVAVTSVFGLDGEDHREGDQRRPKEAAEAHERSREARDVTRQKTGEYTDASREAAQEARDRSRATAQEGRHRRQGEGGQGRGRGHSSATRIWR* |
NA |
False |
100.0 |
97.8 |
52.6 |
3.4 |
100.0 |
50.2 |
100.0 |
100.0 |
||
gene6_1_-10:S:G-A |
gene6 |
gene6_1 |
gene6 |
741 |
True |
False |
False |
MDFPGGSGRRPQQQEPEHLPPMTPLPLARQGSVYSLTFDEFQSSLGGAAKDFGSMNMDELLRSIWSAEEVHSVAAASASAADHAHAAARGPVSIQHQGSLTLPRTLSQKTVDEVWRDLTCVGGGPSSGSAAPAAPPPPAQRHPTLGEITLEEFLVRAGVVREDMTAPPPVPPAPVCPAPAPRPPVLFPHGNVLAPLVPPLQFGNGFVSGAVGQQRGGPVPPAVSPRPVTASAFGKMEGDDLSSLSPSPVPYIFGGGLRGRKPPAMEKVVERRQRRMIKNRESAARSRQRKQKNPHGTGARLNGDGVAVTSVFGLDGEDHREGDQRRPKEAAEAHERSREARDVTRQKTGEYTDASREAAQEARDRSRATAQEGRHRRQGEGGQGRGRGHSSATRIWR* |
100.0 |
False |
NA |
1.1 |
NA |
NA |
NA |
NA |
NA |
NA |
gene6_1_-6:S:T-C |
gene6 |
gene6_1 |
gene6 |
741 |
True |
False |
False |
MDFPGGSGRRPQQQEPEHLPPMTPLPLARQGSVYSLTFDEFQSSLGGAAKDFGSMNMDELLRSIWSAEEVHSVAAASASAADHAHAAARGPVSIQHQGSLTLPRTLSQKTVDEVWRDLTCVGGGPSSGSAAPAAPPPPAQRHPTLGEITLEEFLVRAGVVREDMTAPPPVPPAPVCPAPAPRPPVLFPHGNVLAPLVPPLQFGNGFVSGAVGQQRGGPVPPAVSPRPVTASAFGKMEGDDLSSLSPSPAPYIFGGGLRGRKPPAMEKVVERRQRRMIKNRESAARSRQRKQKNPHGTGARLNGDGVAVTSVFGLDGEDHREGDQRRPKEAAEAHERSREARDVTRQKTGEYTDASREAAQEARDRSRATAQEGRHRRQGEGGQGRGRGHSSATRIWR* |
99.7 |
False |
NA |
1.1 |
NA |
NA |
NA |
NA |
NA |
NA |
gene6_1_0:2D:TCA-T |
gene6 |
gene6_1 |
gene6 |
741 |
True |
False |
False |
MDFPGGSGRRPQQQEPEHLPPMTPLPLARQGSVYSLTFDEFQSSLGGAAKDFGSMNMDELLRSIWSAEEVHSVAAASASAADHAHAAARGPVSIQHQGSLTLPRTLSQKTVDEVWRDLTCVGGGPSSGSAAPAAPPPPAQRHPTLGEITLEEFLVRAGVVREDMTAPPPVPPAPVCPAPAPRPPVLFPHGNVLAPLVPPLQFGNGFVSGAVGQQRGGPVPPAVSPRPVTASAFGKMEGDDLSSLSPSGPVHFRWWVEGKEATGYGEGG* |
65.8 |
True |
NA |
NA |
47.4 |
96.6 |
NA |
49.8 |
NA |
NA |
gene7_1_ref |
gene7 |
gene7_1 |
gene7 |
339 |
False |
MQQDLRNSERRNPEQAHPVMSASSTNSAASPAVSGLDYDDTALTLALPGSSAEPAADRKRAHADHDKPPSPKARAVGWPPVRAYRRNALRDEARLVKVAVDGAPYLRKVDLAAHDGYAALLRALHGMFASCLVAGAGADGAGRIDTAAEYMPTYEDKDGDWMLVGDVPFKMFVDSCKRIRLMKSSEAVNLSPRTSSRQ* |
NA |
False |
100.0 |
100.0 |
41.8 |
53.1 |
49.2 |
51.1 |
44.0 |
44.4 |
||
gene7_1_-1:1D:CG-C |
gene7 |
gene7_1 |
gene7 |
339 |
True |
False |
False |
MQQDLRNSERRNPEQAHPVMSASSTNSAASPAVSGLDYDDTALTLALPGSSAEPAADRKRAHADHDKPPSPKARPWAGRRSARTGATRCATRPGS* |
43.4 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
54.6 |
NA |
gene7_1_2:1I:G-GA |
gene7 |
gene7_1 |
gene7 |
339 |
True |
False |
False |
MQQDLRNSERRNPEQAHPVMSASSTNSAASPAVSGLDYDDTALTLALPGSSAEPAADRKRAHADHDKPPSPKARDRGLAAGPRVPAQRAARRGQAREGGRGRRAVPAEGGPRGARRVRGPAPRAPRHVRLLPRCRSRSRRGGADRHRRRVHAHLRGQGRRLDARRRRPLQDVRGLVQEDPPHEELRGRQPISEDIIPAVIVVGVDAICPTRLISPN* |
49.0 |
True |
NA |
NA |
58.2 |
46.9 |
50.8 |
48.9 |
1.4 |
55.6 |
gene8_1_ref |
gene8 |
gene8_1 |
gene8 |
1825 |
False |
MAFLVERCGEMVVSMESPHAKPVPAPFLTKTYQLVDDPCTDHIVSWGDDDTTFVVWRPPEFARDLLPNYFKHNNFSSFVRQLNTYGFRKIVADRWEFANEFFRKGAKHLLAEIHRRKSSQPLPTPMPPHQPYHHHLHHLHHHLSPFSPPPLAQPVPSYHHHHFQEEPIATATAPHGGAQAGAAGGGNNEGSGAGSRRGLSGRANHVAPVTSPSSAAHASLPSAAGGGAAASSCRLMELDPADSPSPPRRPEADDGTDTVKLFGVALQGKKKKRAHQEDGDDGNHEQGSSDV* |
NA |
False |
100.0 |
98.4 |
55.0 |
65.5 |
54.6 |
52.5 |
63.6 |
51.5 |
||
gene8_1_2:1I:C-CT |
gene8 |
gene8_1 |
gene8 |
1825 |
True |
False |
False |
MAFLVERCGEMVVSMESPHAKPVPAPFLTKTYQLVDDPCTDHIVSWGDDDTTFVVWRPPEFARDLLPNYFKHNNFSSFVRQLNTYGFQEDSGGQVGVRQRVLQEGRQAPTRRDPPEEVVAAAADADAAAPALPPPPPPSPPPPQPVLPAAAGTAGAVVPPPPLPRRAHRHRHRAARRCSSRRRRWRQQ* |
39.1 |
True |
NA |
1.6 |
45.0 |
34.5 |
45.4 |
47.5 |
36.4 |
48.5 |
gene9_1_ref |
gene9 |
gene9_1 |
gene9 |
339 |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVLAIAACELVGGTAAAAVPVACAVEMIHTASLIHDDMPCMDDDALRRGRPSNHVAFGEPTALLAGDALLALAFEHVARGSAGAGVPADRALRAVVELGSVAGVGGIAAGQVADMASEGAPSGSVSLAALEYIHVHKTARLVEAAAVSGAVVGGGGDGEVERVRRYAHFLGLLGQVVDDVLDVTGTSEQLGKTAGKDVAAGKATYPRLMGLKGARAYMGELLAKAEAELDGLDAAPTAPLRHLARGGDHIDVMVVVGYDWVGFGVGIG* |
NA |
False |
100.0 |
100.0 |
49.3 |
55.0 |
53.7 |
54.2 |
29.6 |
38.5 |
||
gene9_1_-2:1D:GC-G |
gene9 |
gene9_1 |
gene9 |
339 |
True |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVWPSPRASSWAGPRPRPSRWRAPSR* |
34.0 |
True |
NA |
NA |
NA |
NA |
46.3 |
NA |
2.3 |
2.4 |
gene9_1_-2:1D:GC-G,-4:1D:TG-T |
gene9 |
gene9_1 |
gene9 |
339 |
True |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVCHRRVRARGRDRGRGRPGGVRRRDDPHRVAHPRRHAVHGRRRAPPRPPLQPRRVRRAHGATRRRRAAGARFRARRPRQRGRRRPRGPRAPRRRGARERSWRRRHRRGAGRRHGERGSPLRLREPGRAGVHPCA* |
38.9 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
NA |
42.1 |
gene9_1_-2:1I:G-GA |
gene9 |
gene9_1 |
gene9 |
339 |
True |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVTGHRRVRARGRDRGRGRPGGVRRRDDPHRVAHPRRHAVHGRRRAPPRPPLQPRRVRRAHGATRRRRAAGARFRARRPRQRGRRRPRGPRAPRRRGARERSWRRRHRRGAGRRHGERGSPLRLREPGRAGVHPCA* |
38.9 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
5.6 |
8.9 |
gene9_1_-2:1I:G-GC |
gene9 |
gene9_1 |
gene9 |
339 |
True |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVPGHRRVRARGRDRGRGRPGGVRRRDDPHRVAHPRRHAVHGRRRAPPRPPLQPRRVRRAHGATRRRRAAGARFRARRPRQRGRRRPRGPRAPRRRGARERSWRRRHRRGAGRRHGERGSPLRLREPGRAGVHPCA* |
38.9 |
True |
NA |
NA |
NA |
NA |
NA |
NA |
1.2 |
3.0 |
gene9_1_-2:1I:G-GT |
gene9 |
gene9_1 |
gene9 |
339 |
True |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVSGHRRVRARGRDRGRGRPGGVRRRDDPHRVAHPRRHAVHGRRRAPPRPPLQPRRVRRAHGATRRRRAAGARFRARRPRQRGRRRPRGPRAPRRRGARERSWRRRHRRGAGRRHGERGSPLRLREPGRAGVHPCA* |
38.9 |
True |
NA |
NA |
50.7 |
45.0 |
NA |
NA |
56.5 |
1.1 |
gene9_1_-2:2D:GCT-G |
gene9 |
gene9_1 |
gene9 |
339 |
True |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVGHRRVRARGRDRGRGRPGGVRRRDDPHRVAHPRRHAVHGRRRAPPRPPLQPRRVRRAHGATRRRRAAGARFRARRPRQRGRRRPRGPRAPRRRGARERSWRRRHRRGAGRRHGERGSPLRLREPGRAGVHPCA* |
38.9 |
True |
NA |
NA |
NA |
NA |
NA |
45.8 |
2.5 |
1.9 |
gene9_1_0:3D:GTGC-G |
gene9 |
gene9_1 |
gene9 |
339 |
True |
False |
False |
MNKLASCFLQHGAPHTQIFKSYHVQRSPSLQLLENRSVSMTRHRAADRAARGTIIDVAVDSGTSFDFESYLSAKARAVHNALDLTLQGLRCPEVLSESMRYSVLAGGKRLRPVAIAACELVGGTAAAAVPVACAVEMIHTASLIHDDMPCMDDDALRRGRPSNHVAFGEPTALLAGDALLALAFEHVARGSAGAGVPADRALRAVVELGSVAGVGGIAAGQVADMASEGAPSGSVSLAALEYIHVHKTARLVEAAAVSGAVVGGGGDGEVERVRRYAHFLGLLGQVVDDVLDVTGTSEQLGKTAGKDVAAGKATYPRLMGLKGARAYMGELLAKAEAELDGLDAAPTAPLRHLARGGDHIDVMVVVGYDWVGFGVGIG* |
99.7 |
False |
NA |
NA |
NA |
NA |
NA |
NA |
2.3 |
2.1 |
Reference |
Locus |
SNP |
INDEL |
Haplotype_Name |
sample1 |
sample2 |
sample3 |
sample4 |
sample5 |
sample6 |
sample7 |
sample8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
gene1 |
gene1_1 |
() |
() |
nan |
0.0 |
0.0 |
0.0 |
0.0 |
0.0 |
0.0 |
0.0 |
0.0 |
gene2 |
gene2_1 |
nan |
(3792, ‘ACT’, ‘A’) |
-1:2D:ACT-A |
0.0 |
0.0 |
51.8 |
45.0 |
0.0 |
0.0 |
0.0 |
0.0 |
gene3 |
gene3_1 |
(3145, ‘A’, ‘C’) |
(3120, ‘C’, ‘CG’),(3121, ‘G’, ‘GT’),(3120, ‘CG’, ‘C’),(3120, ‘CG’, ‘C’),(3120, ‘CGG’, ‘C’),(3120, ‘CGGGGT’, ‘C’),(3118, ‘TACGGGGTG’, ‘T’),(3121, ‘GGGGTGAAGTC’, ‘G’),(3085, ‘TGCTCACTTGCTGACATACCTAGGTACTC’, ‘T’),(3116, ‘CCTACGGGG’, ‘C’) |
-1:1I:C-CG,-2:1I:G-GT,-1:1D:CG-C,-1:1D:CG-C,-26:S:A-C,-1:2D:CGG-C,-1:5D:CGGGGT-C,1:8D:TACGGGGTG-T,-2:10D:GGGGTGAAGTC-G,34:28D:TGCTCACTTGCTGACATACCTAGGTACTC-T,3:8D:CCTACGGGG-C |
0.0 |
0.0 |
50.1 |
100.0 |
95.2 |
100.0 |
91.60000000000001 |
92.0 |
gene4 |
gene4_1 |
nan |
(861, ‘G’, ‘GA’),(861, ‘G’, ‘GT’),(860, ‘TG’, ‘T’) |
2:1I:G-GA,2:1I:G-GT,1:1D:TG-T |
0.0 |
0.0 |
51.7 |
35.2 |
0.0 |
0.0 |
46.4 |
58.099999999999994 |
gene5 |
gene5_1 |
nan |
(1212, ‘C’, ‘CA’),(1212, ‘C’, ‘CT’),(1211, ‘AC’, ‘A’),(1212, ‘CCA’, ‘C’),(1212, ‘CCAAT’, ‘C’),(1211, ‘ACCAAT’, ‘A’),(1211, ‘ACCAATCCCTCCTGCTGGTAAG’, ‘A’) |
-2:1I:C-CA,-2:1I:C-CT,-1:1D:AC-A,-2:2D:CCA-C,-2:4D:CCAAT-C,-1:5D:ACCAAT-A,-1:21D:ACCAATCCCTCCTGCTGGTAAG-A |
0.0 |
0.0 |
61.7 |
0.0 |
56.3 |
57.2 |
91.9 |
50.0 |
gene6 |
gene6_1 |
nan |
(739, ‘TCA’, ‘T’) |
0:2D:TCA-T |
0.0 |
0.0 |
47.4 |
96.6 |
0.0 |
49.8 |
0.0 |
0.0 |
gene7 |
gene7_1 |
nan |
(339, ‘G’, ‘GA’),(336, ‘CG’, ‘C’) |
2:1I:G-GA,-1:1D:CG-C |
0.0 |
0.0 |
58.2 |
46.9 |
50.8 |
48.9 |
56.0 |
55.6 |
gene8 |
gene8_1 |
nan |
(1825, ‘C’, ‘CT’) |
2:1I:C-CT |
0.0 |
1.6 |
45.0 |
34.5 |
45.4 |
47.5 |
36.4 |
48.5 |
gene9 |
gene9_1 |
nan |
(339, ‘G’, ‘GA’),(339, ‘G’, ‘GC’),(339, ‘G’, ‘GT’),(339, ‘GC’, ‘G’),(339, ‘GCT’, ‘G’),(339, ‘GC’, ‘G’),(341, ‘TG’, ‘T’) |
-2:1I:G-GA,-2:1I:G-GC,-2:1I:G-GT,-2:1D:GC-G,-2:2D:GCT-G,-2:1D:GC-G,-4:1D:TG-T |
0.0 |
0.0 |
50.7 |
45.0 |
46.3 |
45.8 |
68.1 |
59.400000000000006 |
Reference |
Locus |
SNP |
INDEL |
Haplotype_Name |
sample1 |
sample2 |
sample3 |
sample4 |
sample5 |
sample6 |
sample7 |
sample8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
gene1 |
gene1_1 |
() |
() |
nan |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
gene2 |
gene2_1 |
nan |
(3792, ‘ACT’, ‘A’) |
-1:2D:ACT-A |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
gene3 |
gene3_1 |
(3145, ‘A’, ‘C’) |
(3120, ‘C’, ‘CG’),(3121, ‘G’, ‘GT’),(3120, ‘CG’, ‘C’),(3120, ‘CG’, ‘C’),(3120, ‘CGG’, ‘C’),(3120, ‘CGGGGT’, ‘C’),(3118, ‘TACGGGGTG’, ‘T’),(3121, ‘GGGGTGAAGTC’, ‘G’),(3085, ‘TGCTCACTTGCTGACATACCTAGGTACTC’, ‘T’),(3116, ‘CCTACGGGG’, ‘C’) |
-1:1I:C-CG,-2:1I:G-GT,-1:1D:CG-C,-1:1D:CG-C,-26:S:A-C,-1:2D:CGG-C,-1:5D:CGGGGT-C,1:8D:TACGGGGTG-T,-2:10D:GGGGTGAAGTC-G,34:28D:TGCTCACTTGCTGACATACCTAGGTACTC-T,3:8D:CCTACGGGG-C |
0 |
0 |
1 |
2 |
2 |
2 |
2 |
2 |
gene4 |
gene4_1 |
nan |
(861, ‘G’, ‘GA’),(861, ‘G’, ‘GT’),(860, ‘TG’, ‘T’) |
2:1I:G-GA,2:1I:G-GT,1:1D:TG-T |
0 |
0 |
1 |
1 |
0 |
0 |
1 |
1 |
gene5 |
gene5_1 |
nan |
(1212, ‘C’, ‘CA’),(1212, ‘C’, ‘CT’),(1211, ‘AC’, ‘A’),(1212, ‘CCA’, ‘C’),(1212, ‘CCAAT’, ‘C’),(1211, ‘ACCAAT’, ‘A’),(1211, ‘ACCAATCCCTCCTGCTGGTAAG’, ‘A’) |
-2:1I:C-CA,-2:1I:C-CT,-1:1D:AC-A,-2:2D:CCA-C,-2:4D:CCAAT-C,-1:5D:ACCAAT-A,-1:21D:ACCAATCCCTCCTGCTGGTAAG-A |
0 |
0 |
1 |
0 |
1 |
1 |
2 |
1 |
gene6 |
gene6_1 |
nan |
(739, ‘TCA’, ‘T’) |
0:2D:TCA-T |
0 |
0 |
1 |
2 |
0 |
1 |
0 |
0 |
gene7 |
gene7_1 |
nan |
(339, ‘G’, ‘GA’),(336, ‘CG’, ‘C’) |
2:1I:G-GA,-1:1D:CG-C |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
gene8 |
gene8_1 |
nan |
(1825, ‘C’, ‘CT’) |
2:1I:C-CT |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |
gene9 |
gene9_1 |
nan |
(339, ‘G’, ‘GA’),(339, ‘G’, ‘GC’),(339, ‘G’, ‘GT’),(339, ‘GC’, ‘G’),(339, ‘GCT’, ‘G’),(339, ‘GC’, ‘G’),(341, ‘TG’, ‘T’) |
-2:1I:G-GA,-2:1I:G-GC,-2:1I:G-GT,-2:1D:GC-G,-2:2D:GCT-G,-2:1D:GC-G,-4:1D:TG-T |
0 |
0 |
1 |
1 |
1 |
1 |
1 |
1 |