Recommendations & Troubleshooting
Recommendations
Minimum read depth filter -c
Accurate haplotype frequency estimation requires a minimum read count which is different between sample type (individuals and Pool-Seq) and ploidy levels.
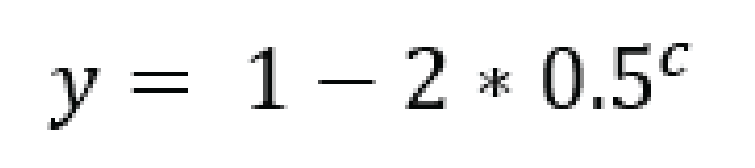

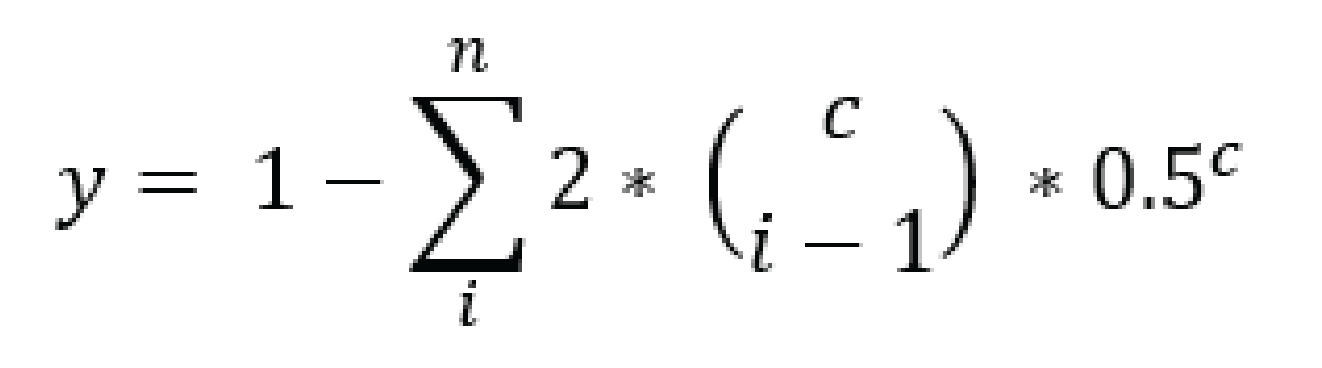
# .. image:: ../images/sites/SMAP_haplotype_diploid_ind_read_count_requirement.png
For tetraploid individuals, calculating the odds of seeing all 4 alleles at least once is a little more complicated than in diploids. A function that approximates this distribution is given by Joly et al. (2006) as
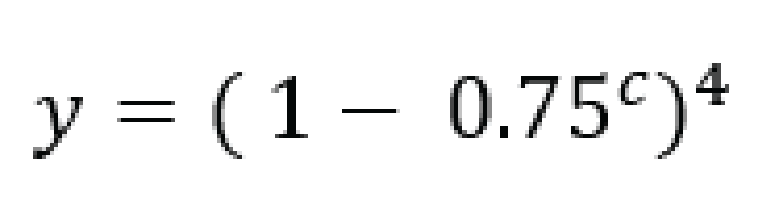
and results in a 95% chance to see all alleles at read count 15 and a 99% chance at around read count 20 (only the full black line should be considered). Figure and additional explanation Griffin et al., 2011. Just like in diploids, in order to see at least 2 copies of each allele it would be best to add a few reads to the results acquired for single copy sightings.
# .. image:: ../images/sites/SMAP_haplotype_tetraploid_ind_read_count_requirement.png
For Pool-Seq data analysis the number of required reads depends on the ploidy as well as the number of samples in a pool, see Raineri et al. (2012), Gautier et al. (2014), and Schlötterer et al. (2014).
Therefore, the user is advised to use the read count threshold to ensure that the reported haplotype frequencies per locus are indeed based on sufficient read data. If a locus has a total haplotype count below the user-defined minimal read count threshold (option -c; default 0, recommended 10 for diploid individuals, 20 for tetraploid individuals, and 30 for pools) then all haplotype observations are removed for that sample.
Troubleshooting
FASTQ Sequence identifier format
sed -i 's|#| |g' *.fq .@ILLUMINA-52179E_0009:8:1:1057:18188#CAGATC/1
ATCGCGGGCAACGGCAGCGCCAGNTAGGGCGGCGCCGGCTACGTTTCCTG
+ILLUMINA-52179E_0009:8:1:1057:18188#CAGATC/1
dcddddcZ`^Lb^bbccddTb^cBTLTbSPL_F_]Y`b_YL]\ILK_\[Z
@ILLUMINA-52179E_0009:8:1:1057:18188 CAGATC/1
ATCGCGGGCAACGGCAGCGCCAGNTAGGGCGGCGCCGGCTACGTTTCCTG
+ILLUMINA-52179E_0009:8:1:1057:18188 CAGATC/1
dcddddcZ`^Lb^bbccddTb^cBTLTbSPL_F_]Y`b_YL]\ILK_\[Z