How It Works
Haplotyping sliding frames with adjacent SNPs
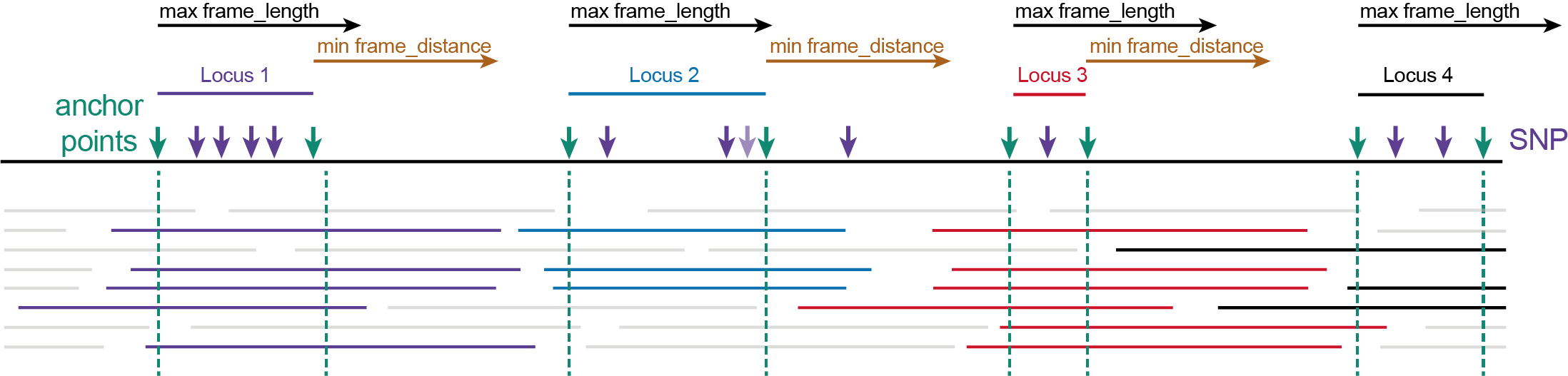
Step 1. Locate the first sliding frame on a reference sequence using a-priori known SNP coordinates (in a sorted VCF file).
 (1) The first SNP on the reference sequence after the off-set length becomes the first SNP of Locus 1 (grey vertical arrow). The frame_distance is not considered before the first locus.(2) The position at distance off-set upstream of the first SNP defines the 5’ start site of Locus 1 (also called the upstream Anchor point, upstream green vertical arrow). If the off-set is set to “0”, the SNP position is the upstream Anchor point of Locus 1.(3) Starting from the 5’ start site of Locus 1, all downstream neighboring SNPs within the maximal frame_length minus off-set length are grouped for Locus 1. (So that the coordinate of the last SNP plus off-set length still falls within the maximum frame_length).(4) The position of the last (most downstream) SNP within the group is determined (blue vertical arrow).(5) The 3’ end site of Locus 1 (also called the downstream Anchor point, downstream green vertical arrow) is positioned at the off-set distance after the last SNP in Locus 1. If the off-set is set to “0”, the last SNP position is the downstream Anchor point of Locus 1.
(1) The first SNP on the reference sequence after the off-set length becomes the first SNP of Locus 1 (grey vertical arrow). The frame_distance is not considered before the first locus.(2) The position at distance off-set upstream of the first SNP defines the 5’ start site of Locus 1 (also called the upstream Anchor point, upstream green vertical arrow). If the off-set is set to “0”, the SNP position is the upstream Anchor point of Locus 1.(3) Starting from the 5’ start site of Locus 1, all downstream neighboring SNPs within the maximal frame_length minus off-set length are grouped for Locus 1. (So that the coordinate of the last SNP plus off-set length still falls within the maximum frame_length).(4) The position of the last (most downstream) SNP within the group is determined (blue vertical arrow).(5) The 3’ end site of Locus 1 (also called the downstream Anchor point, downstream green vertical arrow) is positioned at the off-set distance after the last SNP in Locus 1. If the off-set is set to “0”, the last SNP position is the downstream Anchor point of Locus 1.
Step 2. Locate the next sliding frame at some distance downstream of Locus 1.
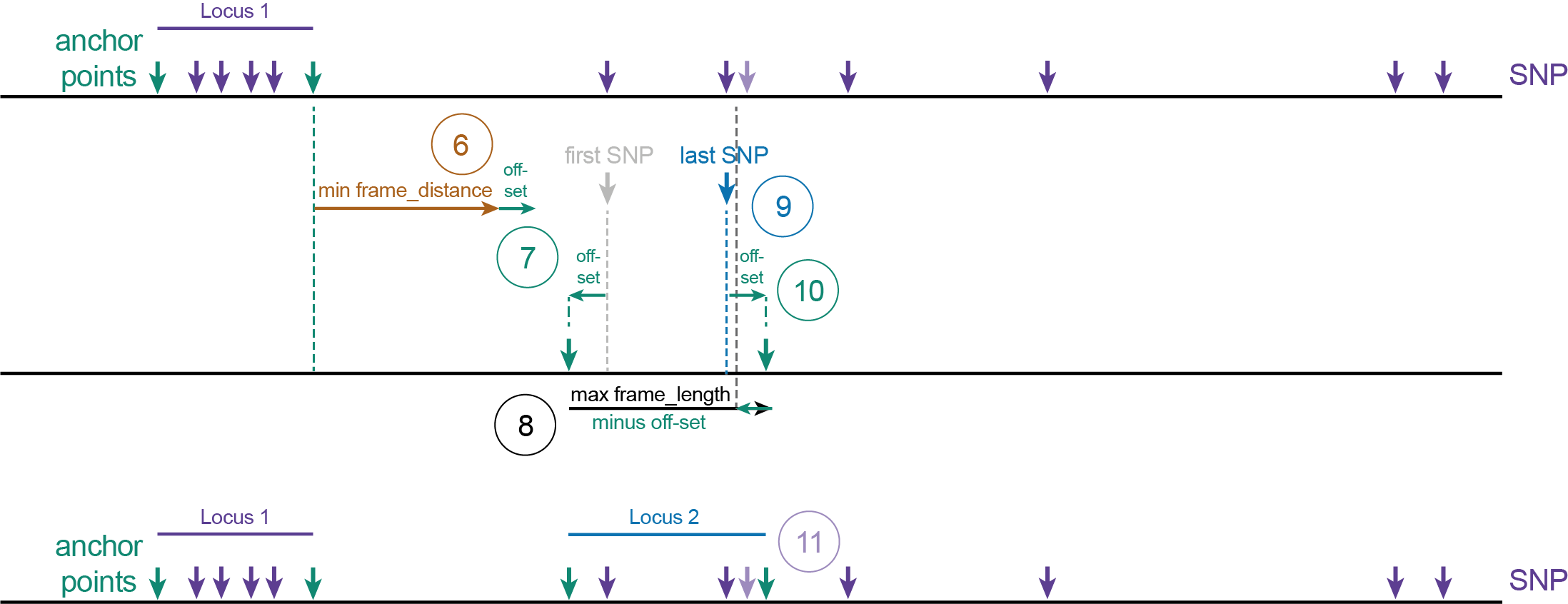 (6) The first SNP after a distance with length minimum frame_distance plus off-set after the 3’ end site of Locus 1, becomes the first SNP of Locus 2.(7) The position at distance off-set upstream of the first SNP defines the 5’ start site of Locus 2.(8) All downstream neighboring SNPs within the frame length minus off-set length are grouped for Locus 2. (So that that SNP plus off-set length still falls within the maximum frame_length).(9) The position of the last SNP within the selected group is determined.(10) The 3’ end site of Locus 2 is positioned at the off-set distance after the last SNP in Locus 2.(11) Note that the SNP (shaded purple vertical arrow) positioned in the off-set region, inbetween the ‘last SNP’ (blue vertical arrow) and the 3’ end site of Locus 2 (downstream green vertical arrow), was not considered to define the locus start and end point coordinates but will still be taken along for haplotyping as it is contained within the Locus 2 range.
(6) The first SNP after a distance with length minimum frame_distance plus off-set after the 3’ end site of Locus 1, becomes the first SNP of Locus 2.(7) The position at distance off-set upstream of the first SNP defines the 5’ start site of Locus 2.(8) All downstream neighboring SNPs within the frame length minus off-set length are grouped for Locus 2. (So that that SNP plus off-set length still falls within the maximum frame_length).(9) The position of the last SNP within the selected group is determined.(10) The 3’ end site of Locus 2 is positioned at the off-set distance after the last SNP in Locus 2.(11) Note that the SNP (shaded purple vertical arrow) positioned in the off-set region, inbetween the ‘last SNP’ (blue vertical arrow) and the 3’ end site of Locus 2 (downstream green vertical arrow), was not considered to define the locus start and end point coordinates but will still be taken along for haplotyping as it is contained within the Locus 2 range.
Step 3. Repeat along the length of the reference sequence, while ignoring SNPs that are too close to the previous locus.
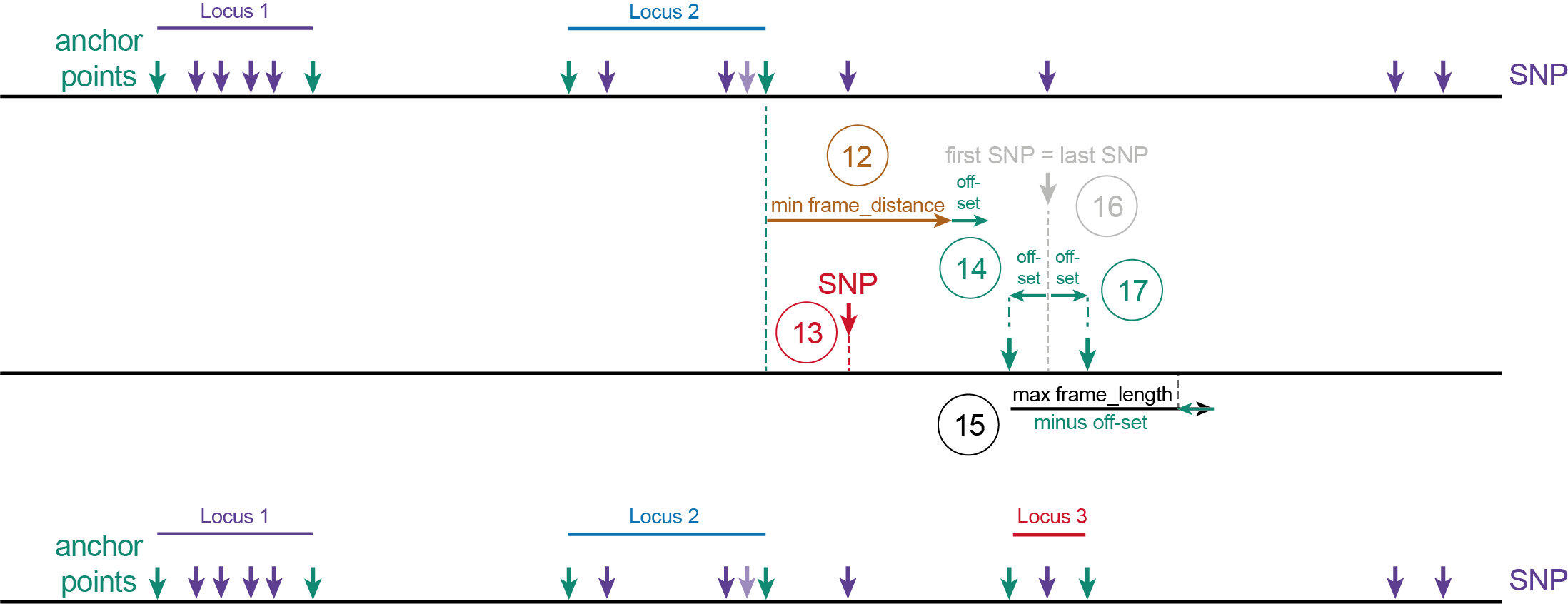 (12) The first SNP after length minimum frame_distance plus off-set after the 3’ end site of Locus 2 becomes the first SNP of Locus 3.(13) SNPs positioned within the frame-distance regions are ignored.(14) The position at distance off-set upstream of the first SNP defines the 5’ start site of Locus 3.(15) All downstream neighboring SNPs within the frame_length minus off-set length are grouped for Locus 3.(16) If only one SNP exists, this also becomes the last SNP.(17) The 3’ end site of Locus 3 is positioned at the off-set distance after the last SNP in Locus 3.
(12) The first SNP after length minimum frame_distance plus off-set after the 3’ end site of Locus 2 becomes the first SNP of Locus 3.(13) SNPs positioned within the frame-distance regions are ignored.(14) The position at distance off-set upstream of the first SNP defines the 5’ start site of Locus 3.(15) All downstream neighboring SNPs within the frame_length minus off-set length are grouped for Locus 3.(16) If only one SNP exists, this also becomes the last SNP.(17) The 3’ end site of Locus 3 is positioned at the off-set distance after the last SNP in Locus 3.
Step 4. Locate the last sliding frame on the reference sequence.
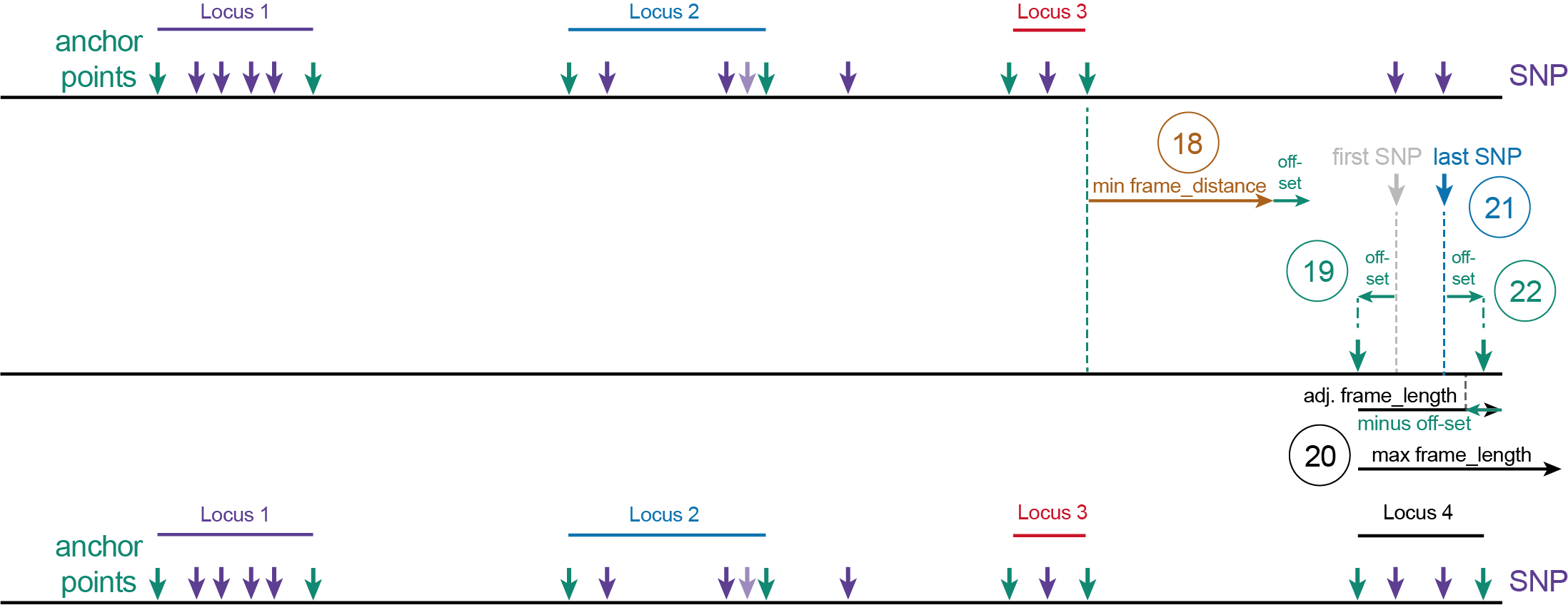 (18) The first SNP after length minimum frame_distance plus off-set after the 3’ end site of Locus 3 becomes the first SNP of Locus 4.(19) The position at distance off-set upstream of the first SNP defines the 5’ start site of Locus 4.(20) If the frame_length exceeds the remaining length of the reference sequence, it is set at the last nucleotide of the reference sequence. All downstream neighboring SNPs within the frame_length minus off-set length are grouped for Locus 4.(21) The position of the last SNP within the group is determined for Locus 4. The last SNP can be positioned at maximal the length of the reference sequence minus the off-set length.(22) The 3’ end site of Locus 4 is positioned at the off-set distance after the last SNP.
(18) The first SNP after length minimum frame_distance plus off-set after the 3’ end site of Locus 3 becomes the first SNP of Locus 4.(19) The position at distance off-set upstream of the first SNP defines the 5’ start site of Locus 4.(20) If the frame_length exceeds the remaining length of the reference sequence, it is set at the last nucleotide of the reference sequence. All downstream neighboring SNPs within the frame_length minus off-set length are grouped for Locus 4.(21) The position of the last SNP within the group is determined for Locus 4. The last SNP can be positioned at maximal the length of the reference sequence minus the off-set length.(22) The 3’ end site of Locus 4 is positioned at the off-set distance after the last SNP.
Step 5. Continue the process for all other reference sequences.
Step 6. Use the sliding frames to delineate loci for read-backed haplotyping with SMAP haplotype-sites.
Special cases and the optimal use of parameter settings
According to the following rationale, parameter settings can be optimized to cover special cases.Off-set distances are used to ensure that the sequence context around the SNPs are also covered by the same read.In this case, the outer 5’ and 3’ positions delineating the locus are used as ‘Anchor points’ rather than as polymorphic SNPs and are used for evaluation of complete coverage of the read across the locus length.Always use option--partial excludefor SMAP haplotype-sites.If the off-set is set to “0”, the 5’ end site corresponds to the first SNP, and the 3’ end site of the locus corresponds to the last SNP.If only one SNP exists within the maximal frame_length and off-set is set to “0”, then the locus is limited to length 1 and only the single SNP is scored as haplotype.If only one SNP exists within the maximal frame_length and off-set is set greater than “0”, then the locus is defined by length 1 + 2 x off-set and both the single SNP and the two Anchor points are scored as haplotype.SNPs positioned in the frame-distance regions are ignored.If the frame_distance is set to “0”, loci may become directly adjacent, but frames never overlap.The minimal frame_distance is always respected.Frame_length must always be set at a value greater than or equal to 1 + 2 x off-set.Frame_length must always be set at a value shorter than the longest read length (ideally about one-half to two-thirds). Otherwise, reads can never entirely span the longest frame_length and are dropped by SMAP haplotype-sites.Frame_length is a measure for the maximum length per locus, but the effective locus length distribution is likely smaller and depends on SNP density combined with off-set and frame_length.
Haplotyping the junction sites of large structural variants such as deletions and inversions
Each junction is considered as its own sliding frame
Delineating sliding frames for this application is very simple as all parameters should be fixed.
Locus: name of the region of the reference genome that contains the junction.
Anchor points: the start and end positions of the locus are defined as the nucleotides immediately adjacent to the junction.
maximal frame_length is set to “3”. Each junction is considered separately, the central nucleotide is at the junction.
minimal frame_distance is set to “0”.
off-set is set to “1”: the nucleotides immediately upstream and downstream of the junction are Anchor points by definition.
always use option
--partial includefor SMAP haplotype-sites.