Feature Description
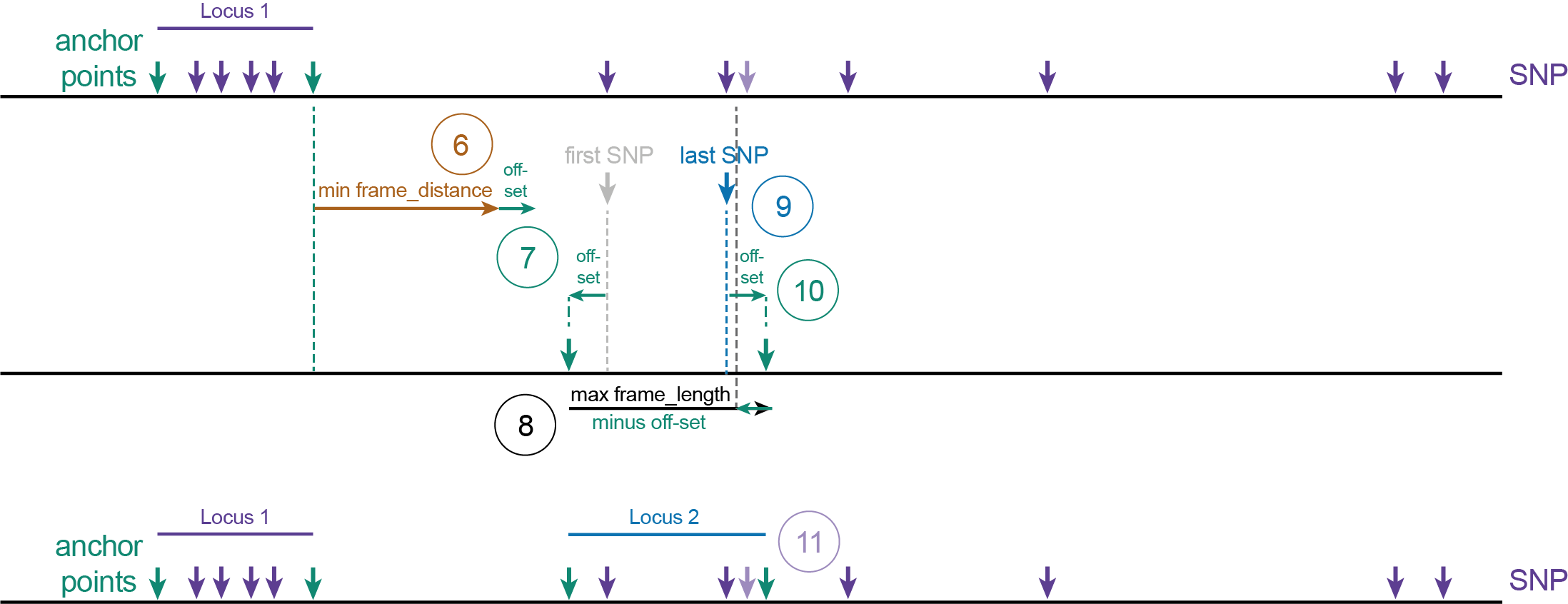
The scheme below defines the features of sliding frames and shows how parameters can be adjusted to customise the length and spacing of sliding frames with respect to SNPs on a given reference genome sequence. Key features are:
Locus: name of the region of the reference genome that contains polymorphisms to be haplotyped.
Anchor points: the start and end positions of the locus.
Maximal frame_length: the maximal length of the frame that includes the first and last SNPs to be grouped, as well as the off-set at start and end of the frame.
Minimal frame_distance: minimal distance between two adjacent loci.
Off-set: a number of nucleotides before the first SNP, and after the last SNP. Used to create space around the SNPs to ensure consistent read mapping around the SNPs to be haplotyped.
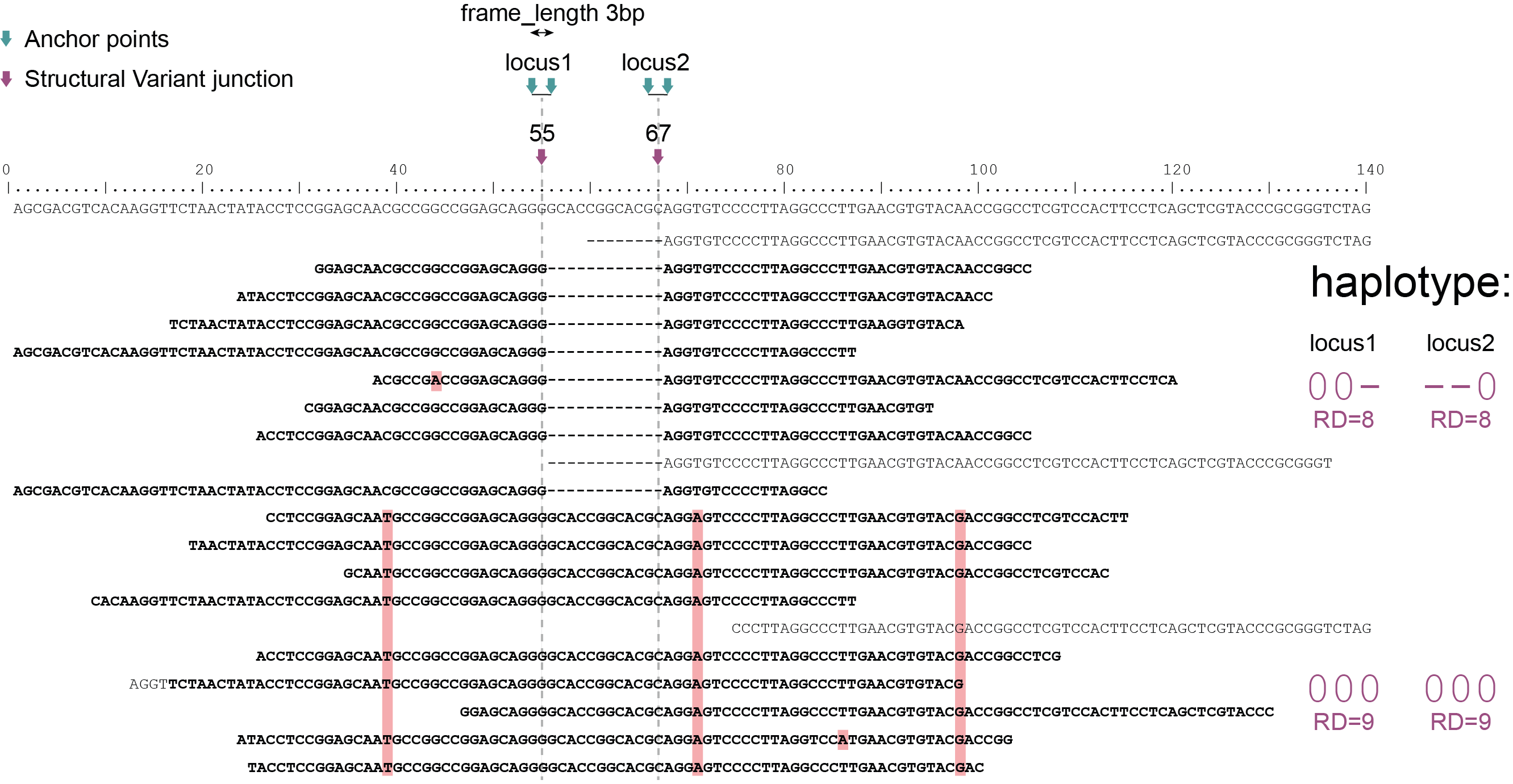
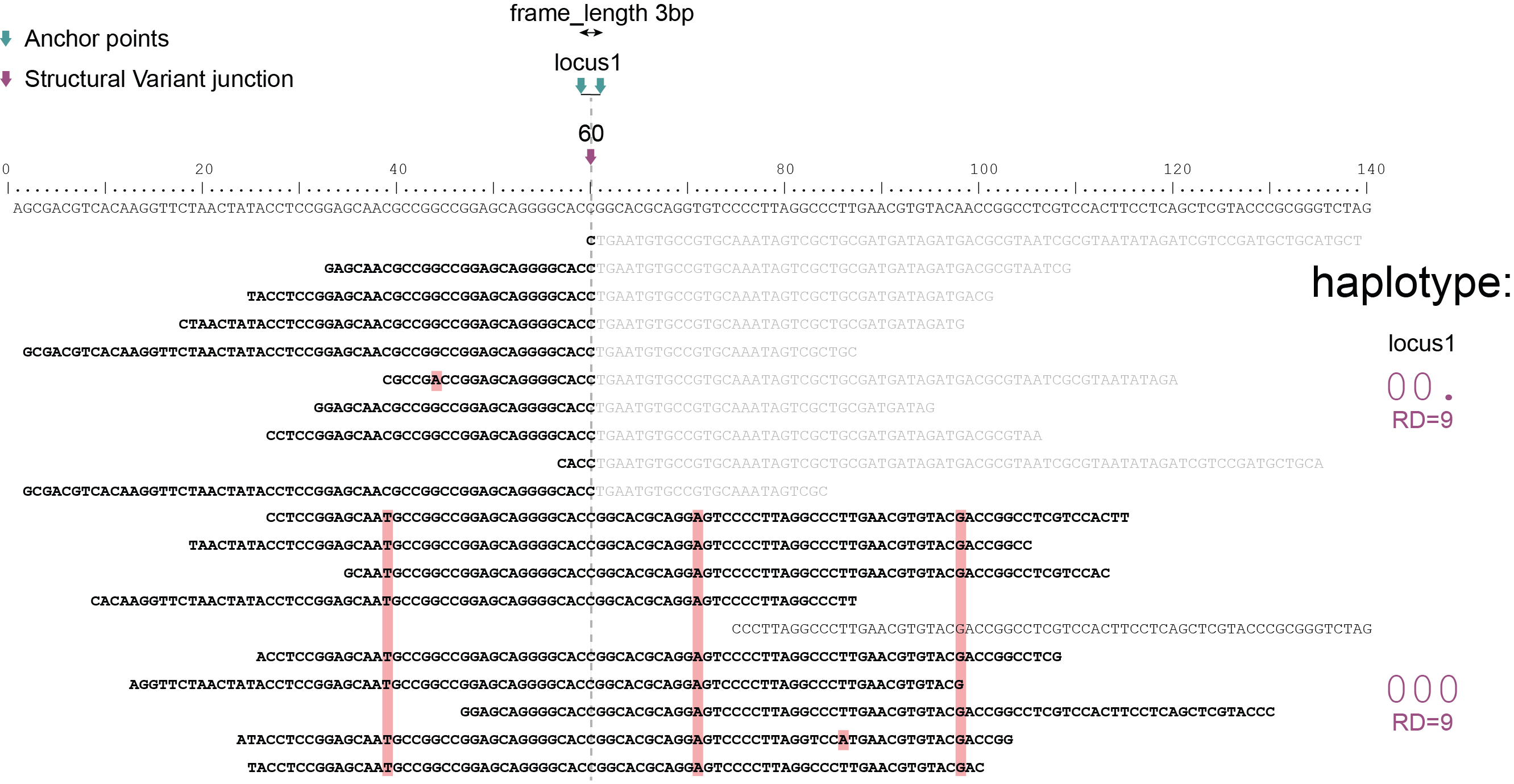
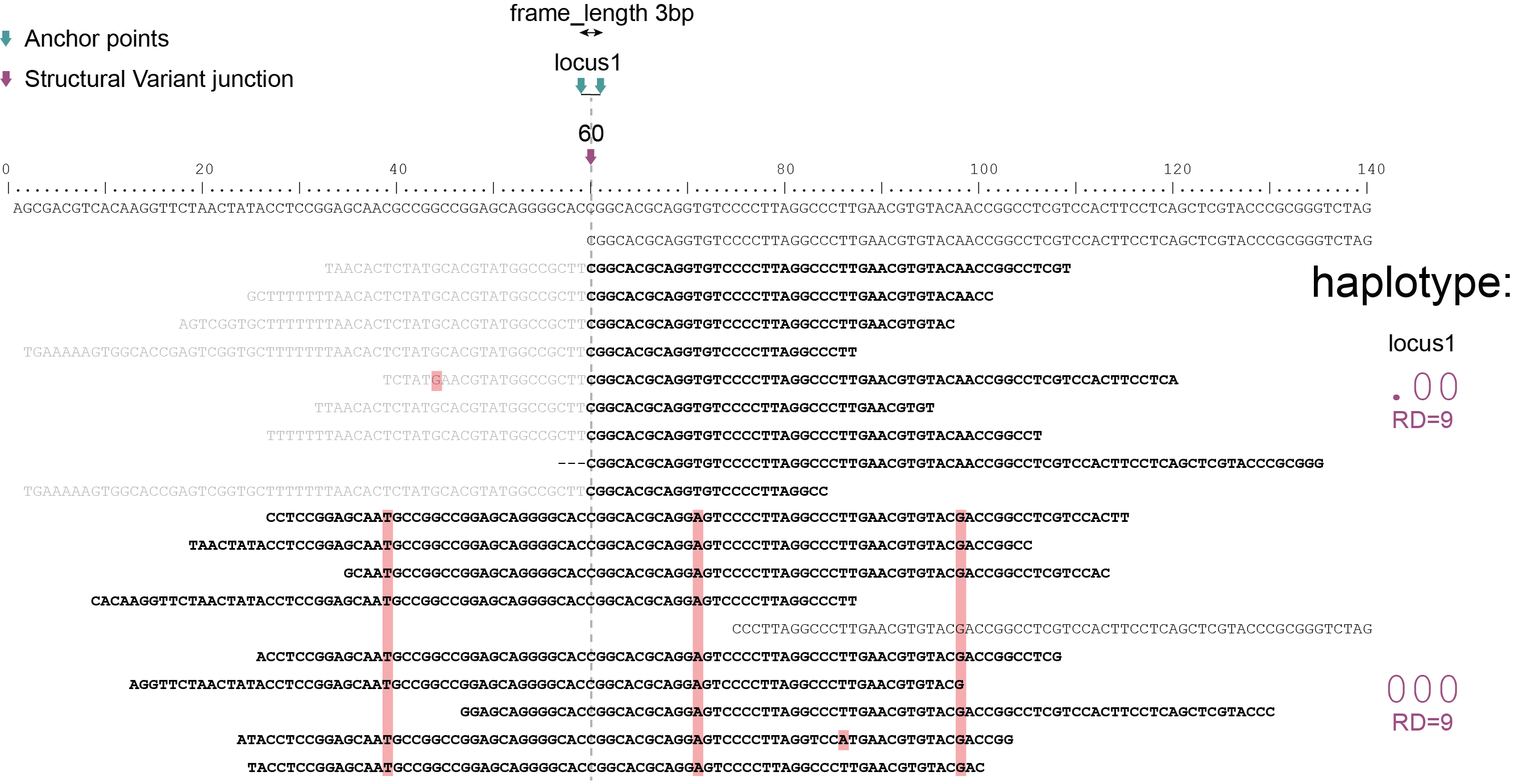
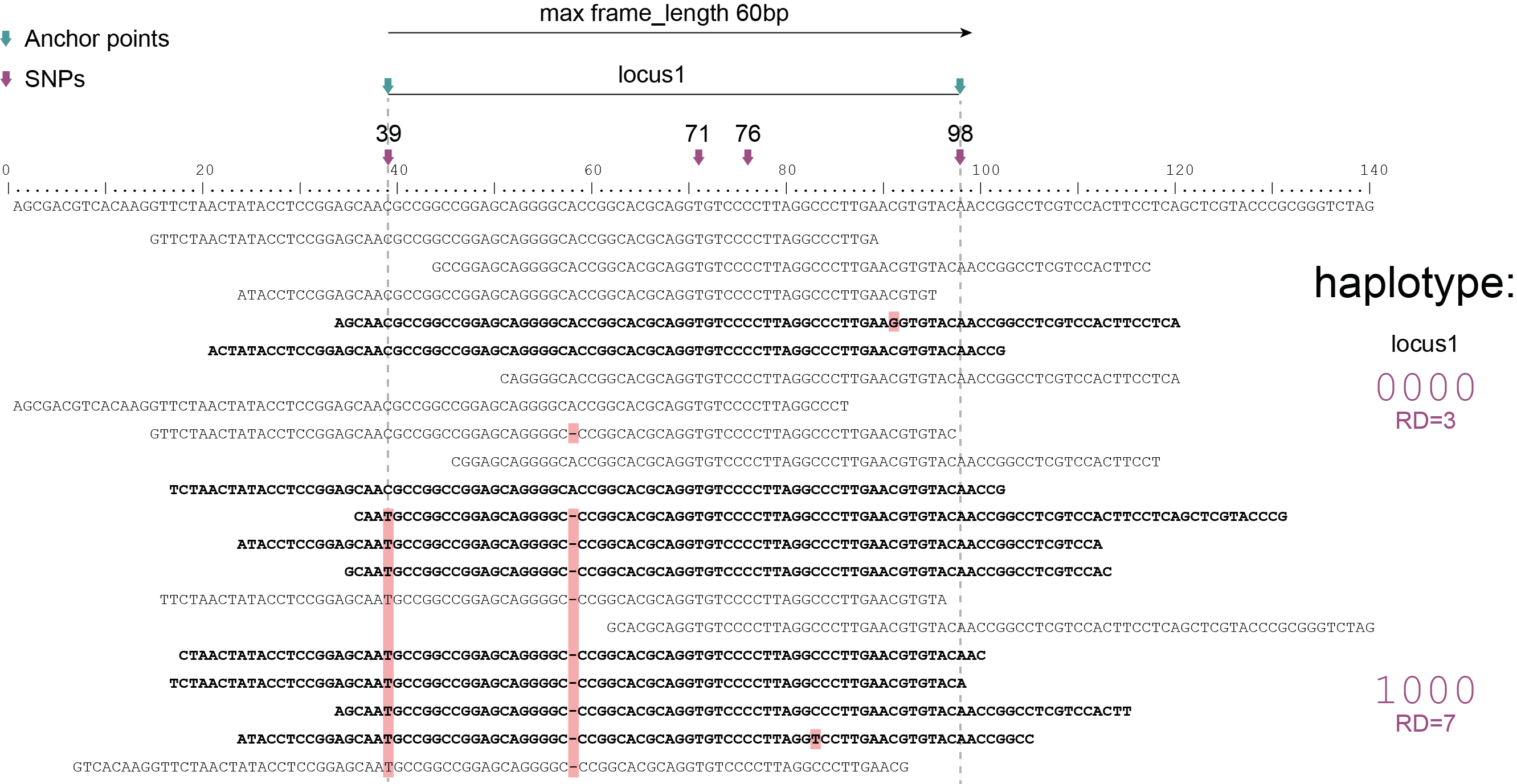
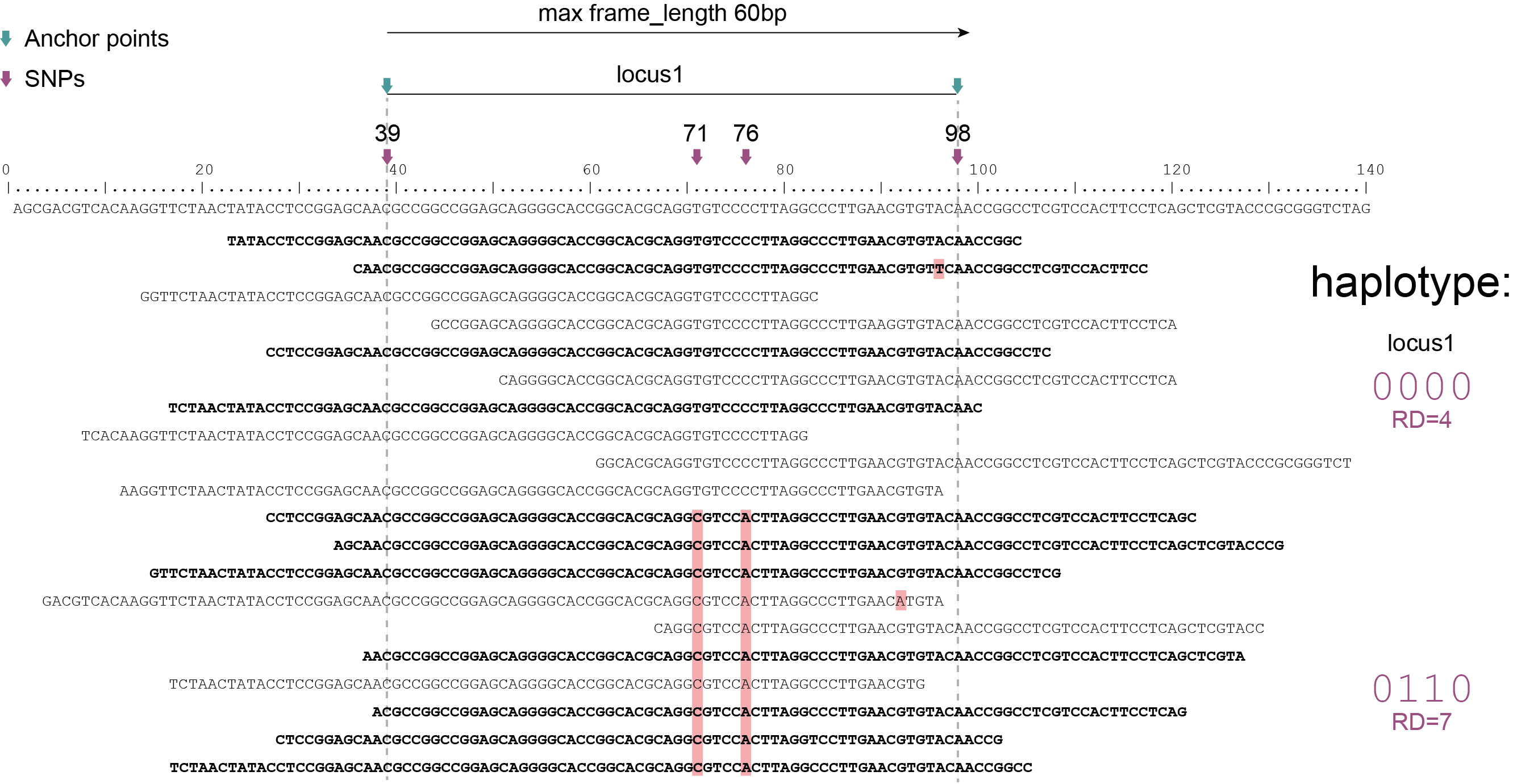
Haplotyping neighboring SNPs in sliding frames
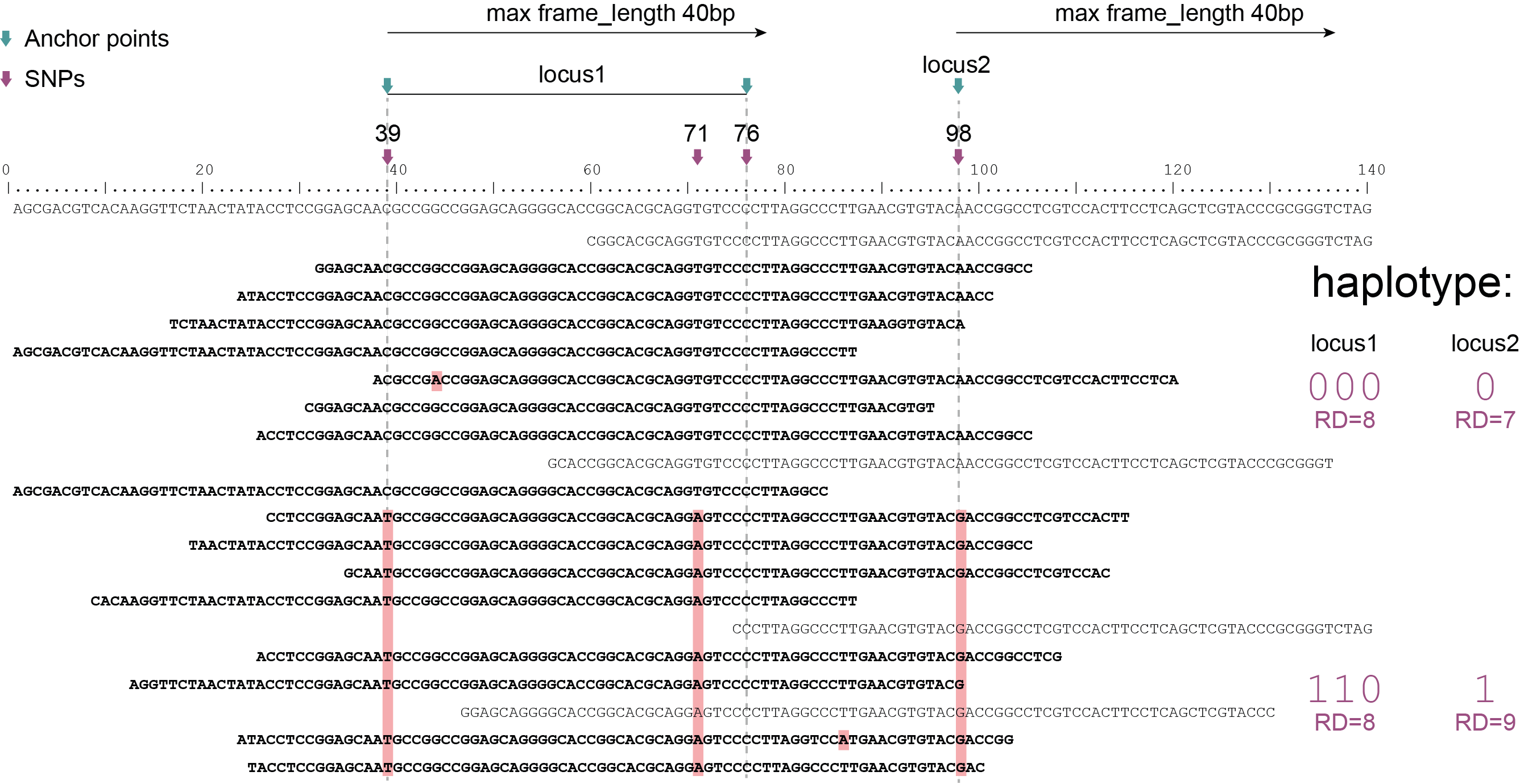
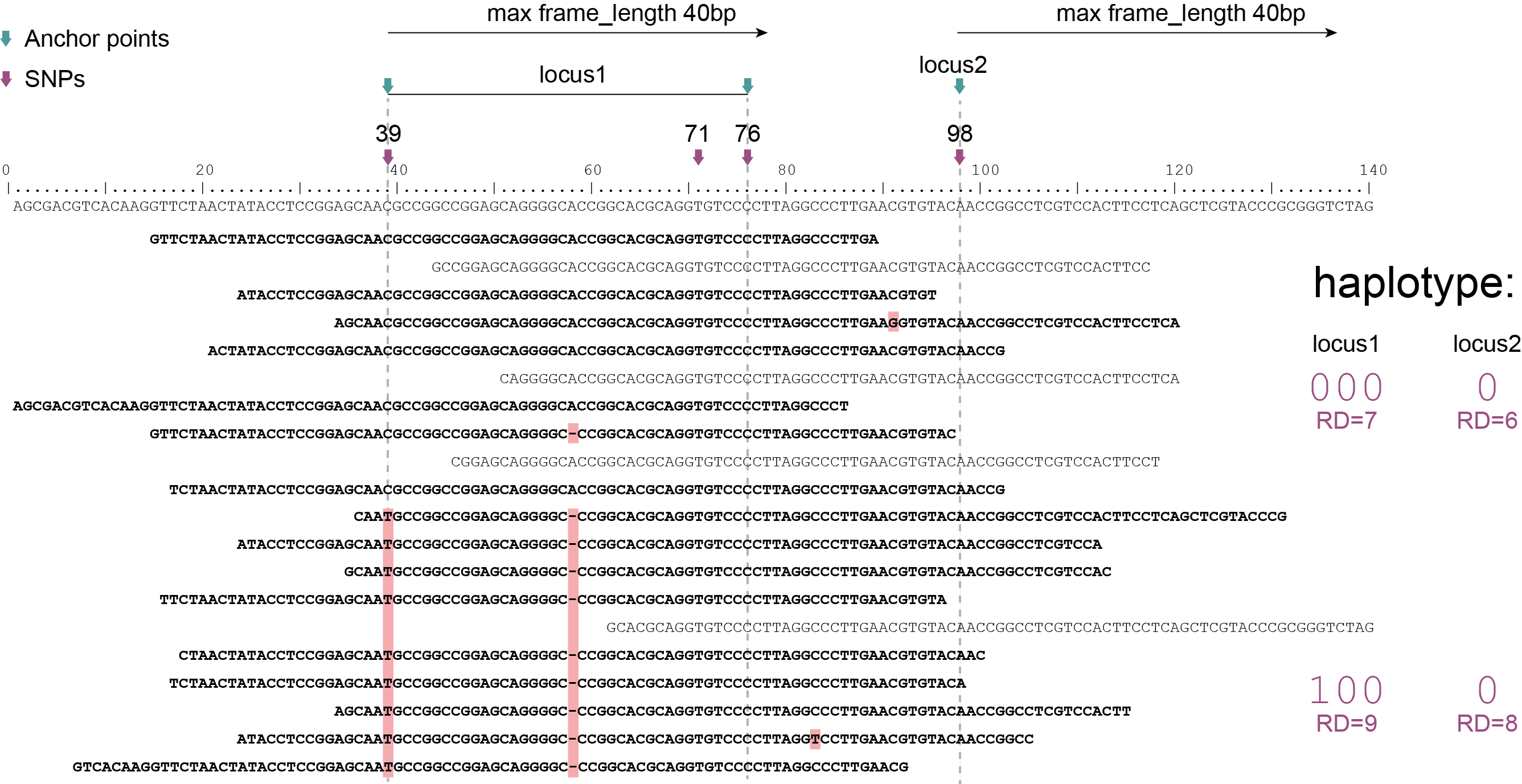
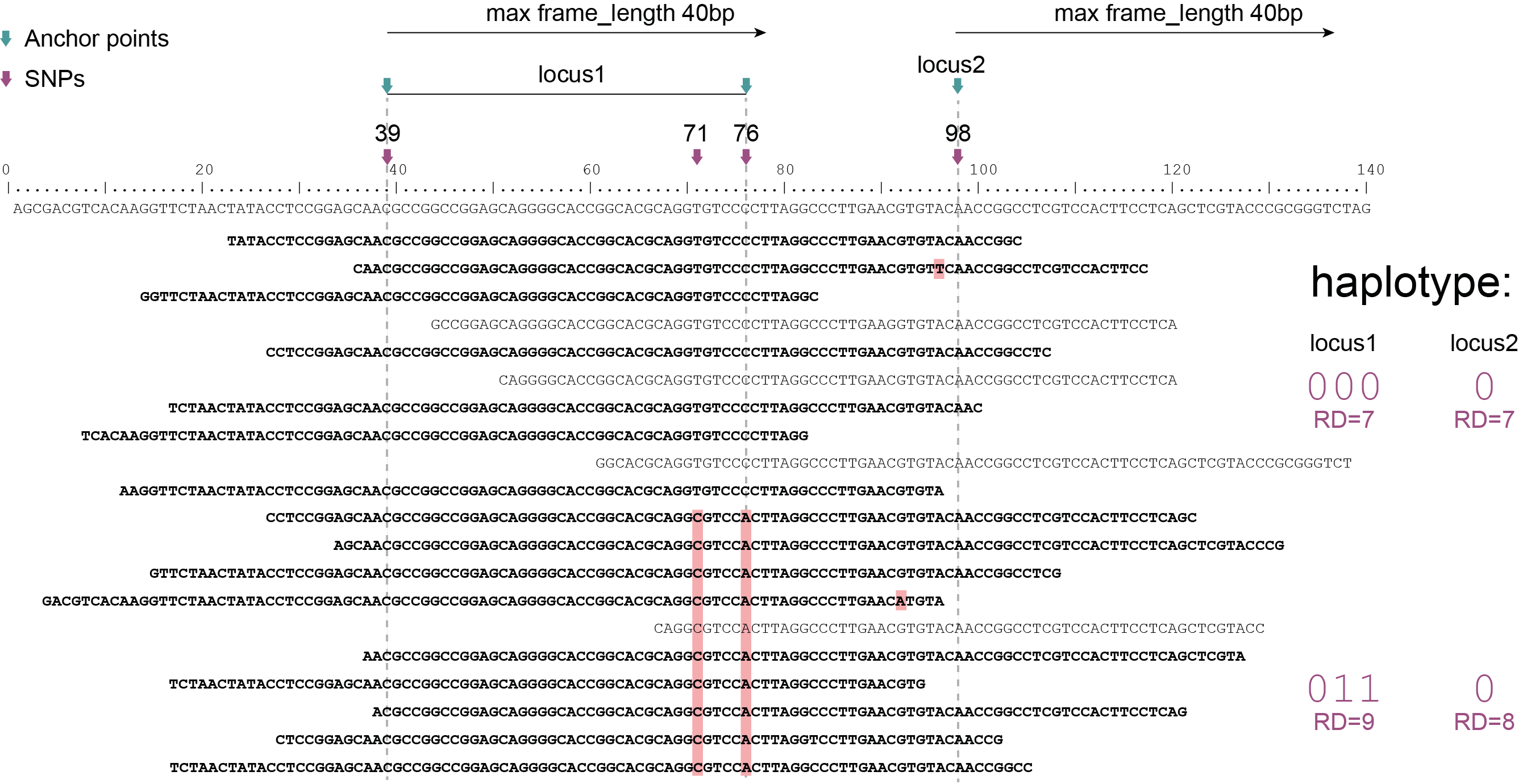
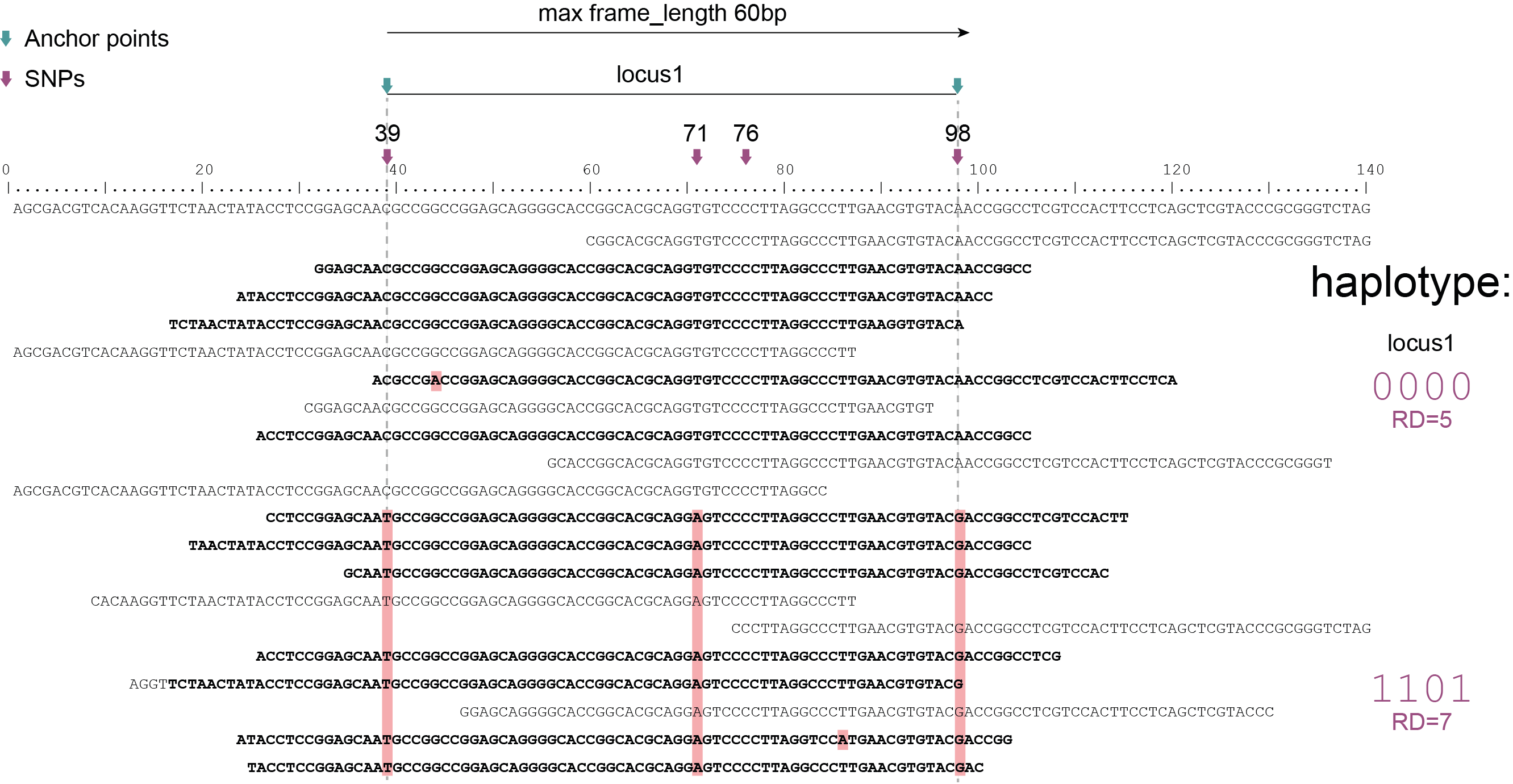
Schemes show a heterozygous individual with a reference allele and one alternative allele. Only reads spanning the entire locus are used for read-backed haplotyping. The total read depth (RD) is shown below each derived haplotype (string of “0“‘s and “1“‘s). Note that at longer sliding frame length (40bp versus 60bp), the number of loci reduces (locus_1 and locus_2 are joined into the longer locus_1), the number of SNPs grouped per locus increases, and the number of reads spanning the entire locus_1 (shown in bold) is reduced so that effective read depth per haplotype (RD) reduces. At shorter maximal sliding frame length (e.g. 40 bp) some reads span both locus_1 and locus_2. Because loci are haplotyped independently, different parts of the same read may add data to neighboring loci, as long as they span the entire length per locus.
Neighboring SNPs define sliding frames. Given a set of SNPs in a VCF file, loci are delineated that contain subsets of neighboring SNPs within a given maximal sliding frame length (e.g. 40bp or 60bp).
Haplotyping junctions of (large-scale) inversions or deletions
The junctions surrounding inversions or deletions can be recognized at the single read level as a sudden breakpoint in the read-reference alignment. Typically, the maximum exact match (MEM) that seeds the alignment in BWA-MEM places the longest half of the read adjacent to the breakpoint, and the other half of the read is soft-clipped, or not matched to the reference genome. At the locus level, it can be recognized as the consistent sharp drop in read depth on one side of the junction, as each read is expected to display the same read-reference alignment breakpoint. Heterozygous individuals are expected to display a pattern where half the number of reads display the alignment breakpoint at the junction, and the other half of the number of reads display continuous read-reference alignments across the junction. This read mapping profile can be coded as haplotype by SMAP, because read-reference alignments are transformed to haplotypes while considering absence/presence of read mapping. The approach to score clean drops in read depth at SV mapping breakpoints is to define 3-bp loci with the breakpoint nucleotide as the central position, immediately flanked by an upstream and a downstream nucleotide position and score absence/presence per position. Deletions with respect to the reference are marked as “-” characters and absence of read mapping (due to terminated read alignment) as “.” characters in the haplotype string.
Please note that the above description refers to large scale inversions (which, in a short read, behave as a ‘deletion’ of the sequence neighboring a junction), not insertions. SMAP haplotype-sites currently does not support identifying insertions (and coding those as haplotype strings), as we strictly adhere to the reference coordinate system to encode the absence or presence of nucleotides in the alignment. Adding nucleotides into the haplotype string (insertions with respect to the reference haplotype string) is not possible.