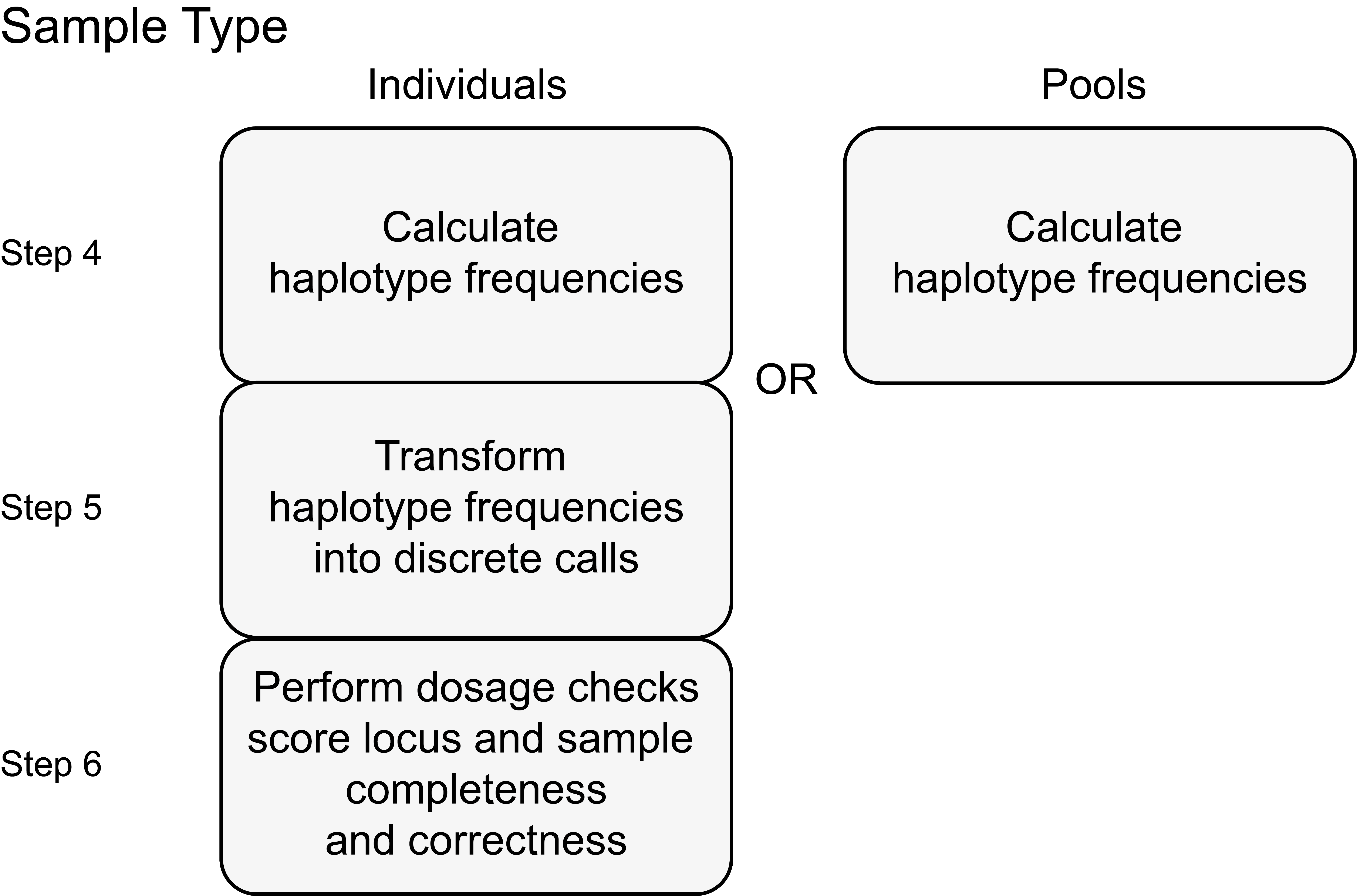
Haplotyping for different sample types (individual or Pool-Seq)
Individual and Pool-Seq samples are analyzed differently in three important ways;
In individuals, low frequency haplotypes (<5%) are generally noise and need to be removed from the data. In Pool-Seq, it may be difficult to discriminate between noise and low frequency true alleles.
In Pool-Seq, haplotype frequencies represent the genetic diversity of the population. For individuals, haplotype frequencies should be converted into discrete genotype calls, which can either be expressed as dominant (0/1) or dosage (0/1/2, diploid; 0/1/2/3/4, tetraploid).
In individuals, the total number of haplotypes (sum of dosage calls per locus per sample) can be compared to the number of alleles present in the nuclear genome (2 for diploids, 4 for tetraploids). This provides to opportunity to remove loci with consistent bad calls across a large fraction of the sample set, or to identify samples with substantial numbers of loci with unexpected dosage calls (thus identifying samples with mis-labeled ploidy levels, or contaminated DNA samples).
Contents: