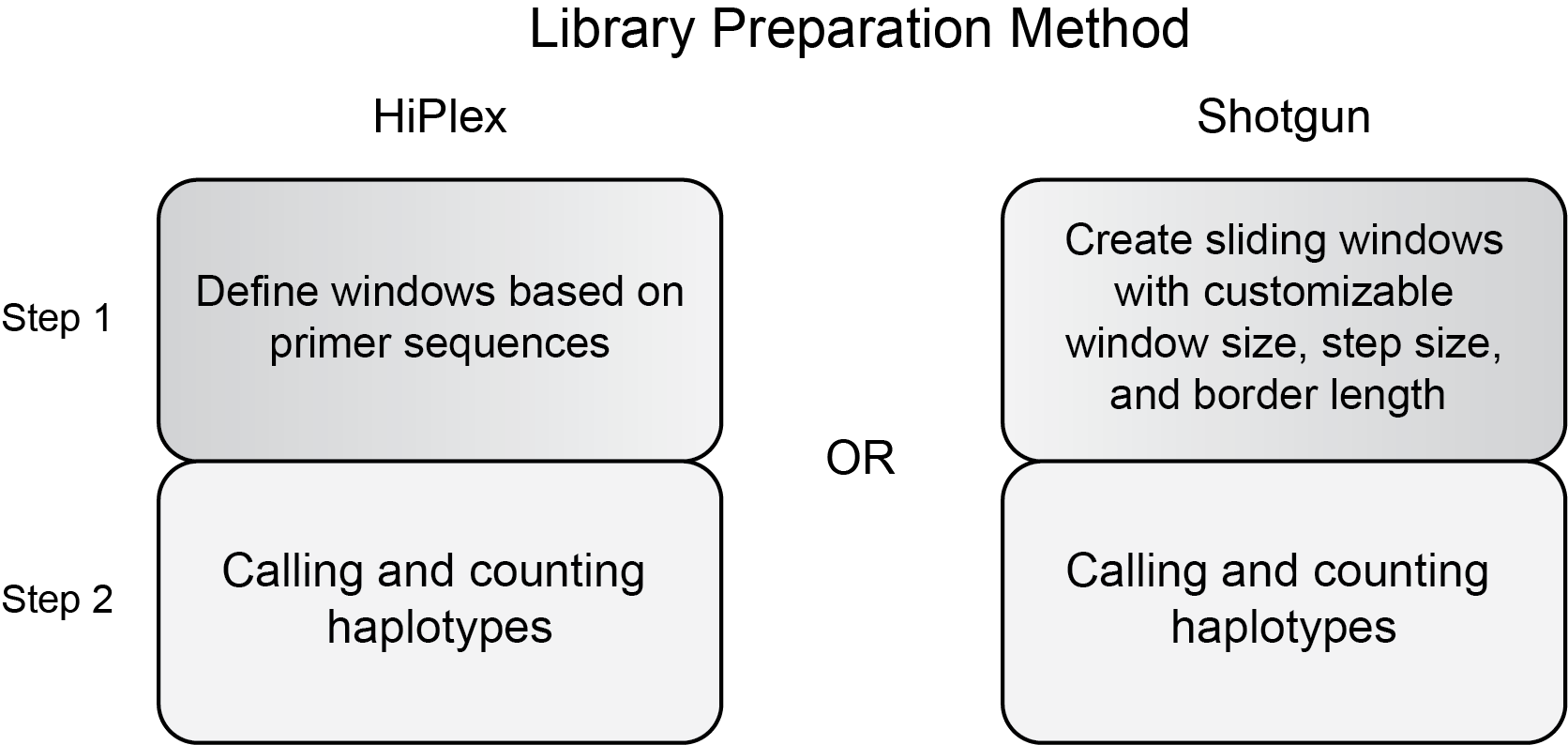
Haplotyping for different library preparation methods
Delineation of Windows for HiPlex data
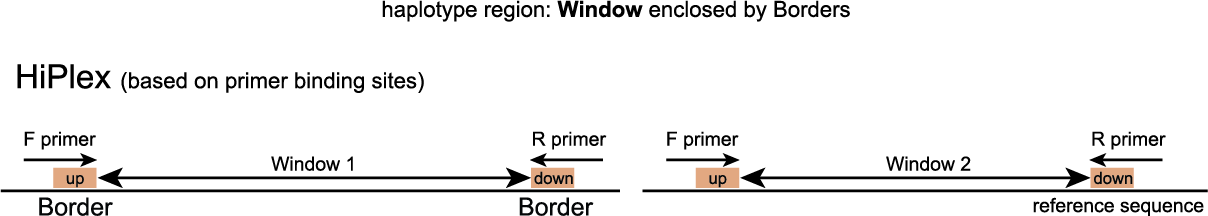
For HiPlex, a simple GFF file defining Windows is created by listing two reference sequence positions (primer-based anchor points) for each border for each amplicon (an upstream border sequence within the forward primer, and a downstream border sequence within the reverse primer). By design, all primer binding sites and expected amplicons are a priori known, and there is no need to search for alternative loci with mapped reads.
Delineation of Windows for Shotgun Sequencing data
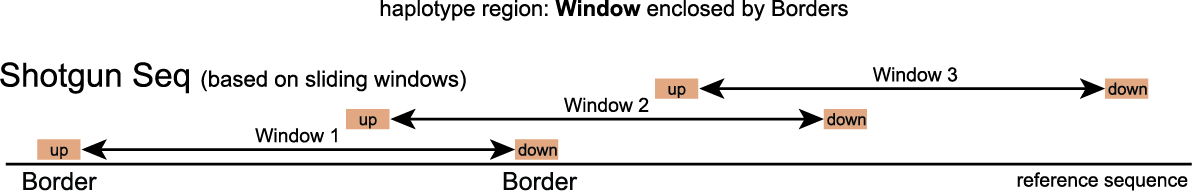
For Shotgun Sequencing, contrary to HiPlex read data, read positions are scattered across the genome and are not “stacked”. Therefore, the delineation of Windows is a little different than in HiPlex data and the concept of sliding windows is introduced. A GFF file containing sliding windows is created by iterating over the reference sequence with a fixed Stepsize and Windowsize. More information can be found on the Shotgun Sequencing page.
If only specific loci are of interest, it is also possible to follow the HiPlex Window delineation procedure.
Haplotype-window considers different locus structures for HiPlex and Shotgun Seq
Contents: