HiPlex amplicon sequencing
Setting the stage
Core
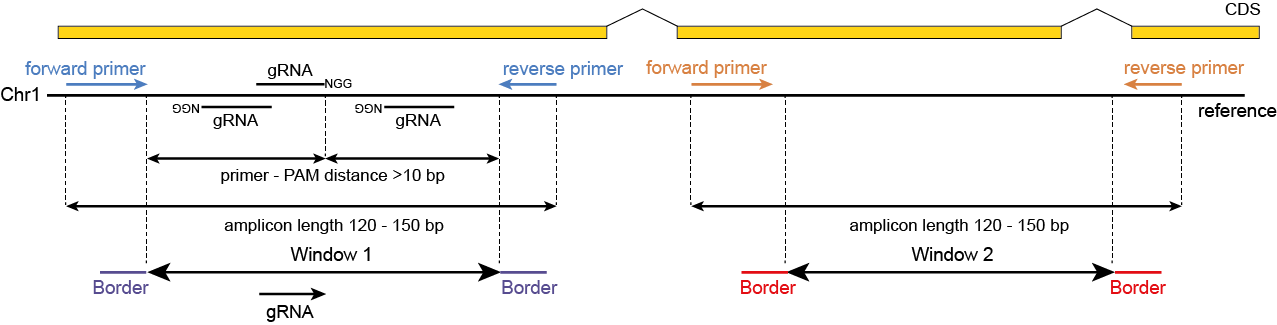
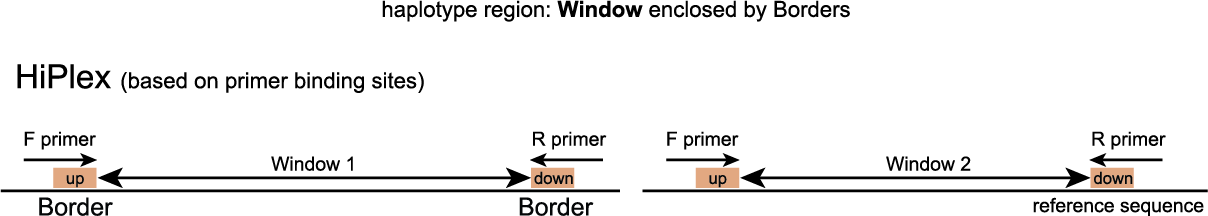
Windows are defined as any region enclosed by a pair of Borders. SMAP haplotype-window considers the entire read sequence spanning the region between the Borders as haplotypes. Any pair of Borders can be chosen and searched for in a given set of reads. Because the primer sequence itself becomes incorporated into the amplicon molecule, (parts of) primers can naturally function as Border sequences delineating the enclosed amplified region.
HiPlex library preparation and preprocessing
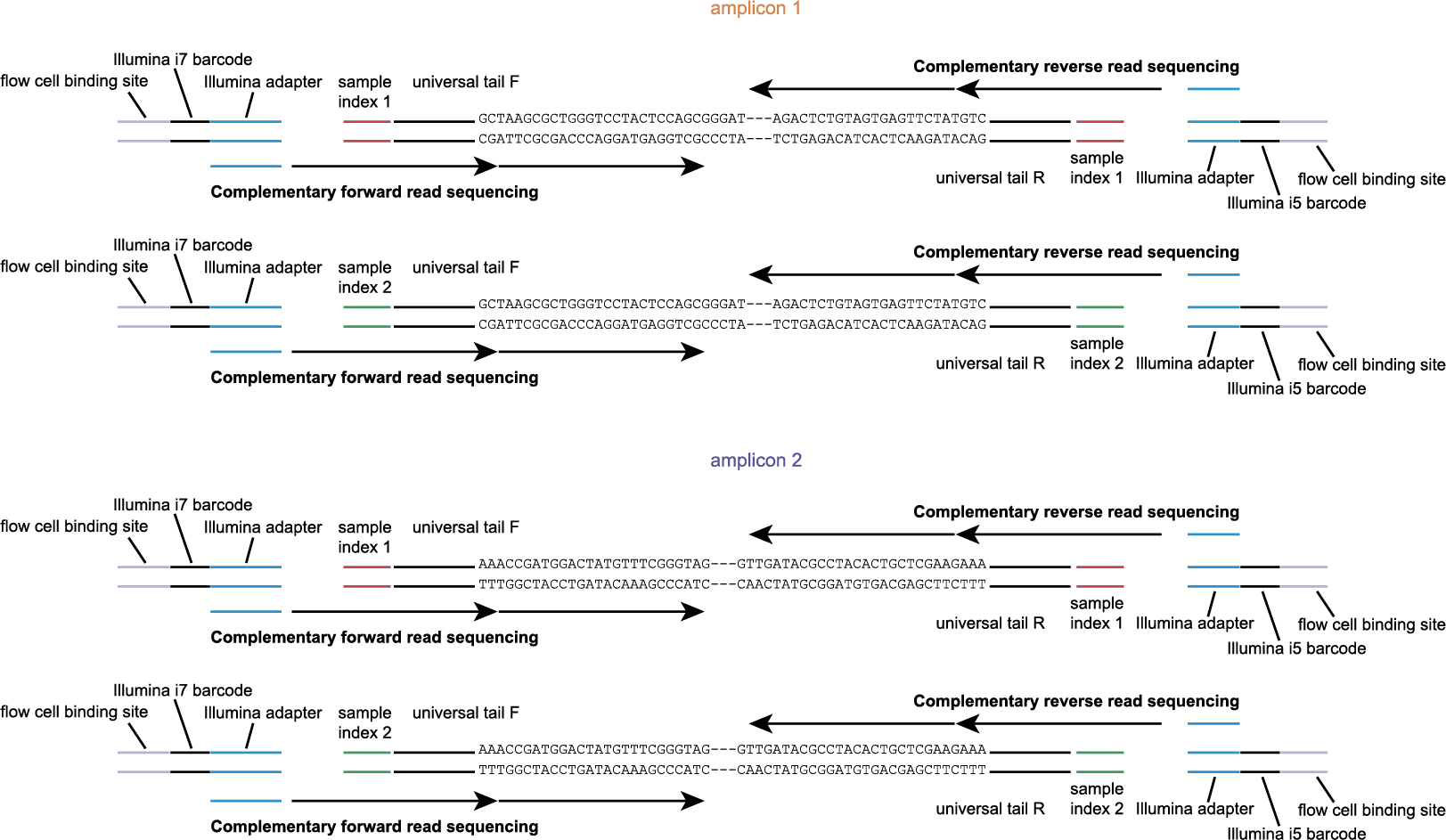
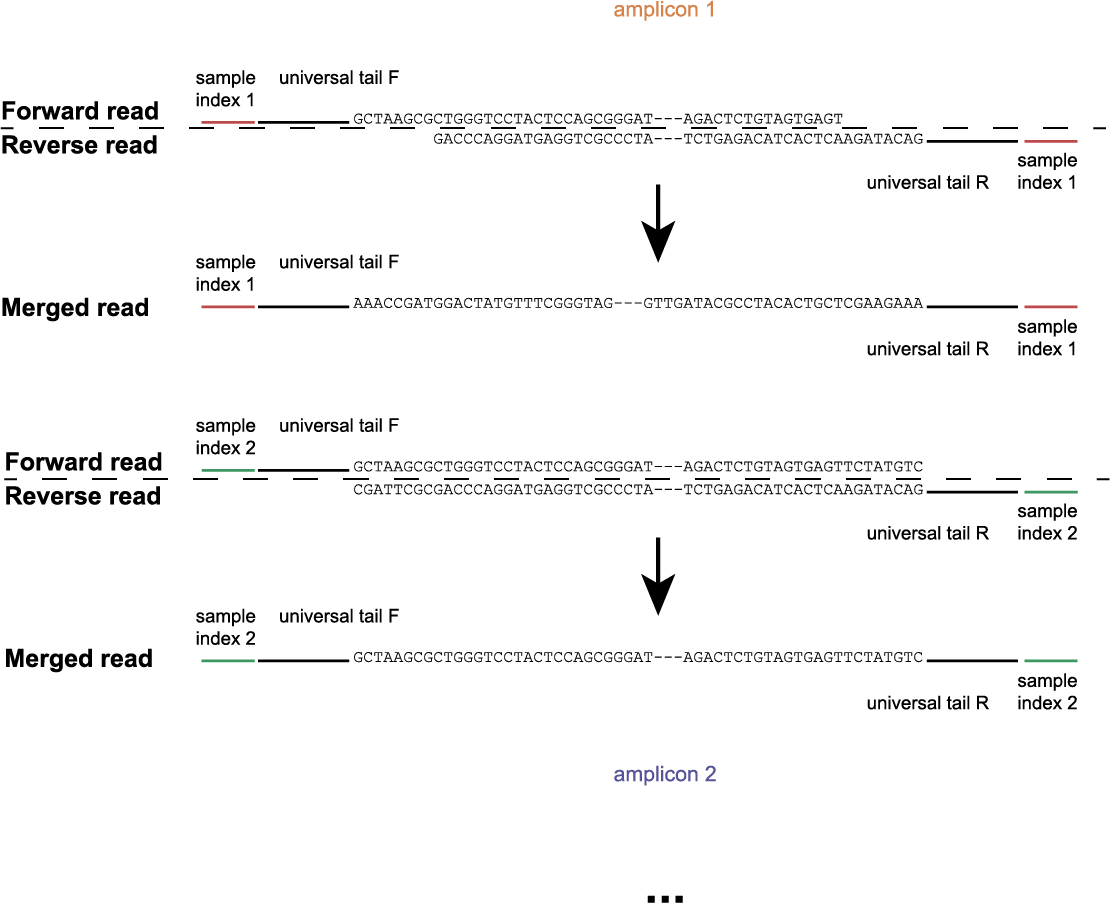
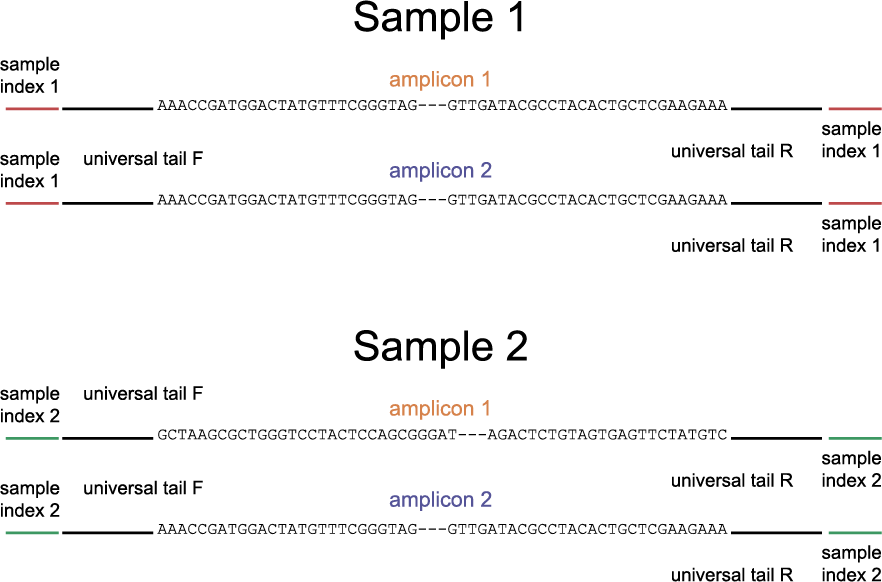
The schemes below show how HiPlex libraries are prepared, sequenced, and trimmed to remove sample-specific barcodes and adapter sequences. The reads obtained by HiPlex amplicon sequencing can be mapped directly onto a reference sequence. Optionally, gRNAs can be designed together with amplicons, so that HiPlex targetted resequencing can be used to characterize CRISPR/Cas induced mutations.
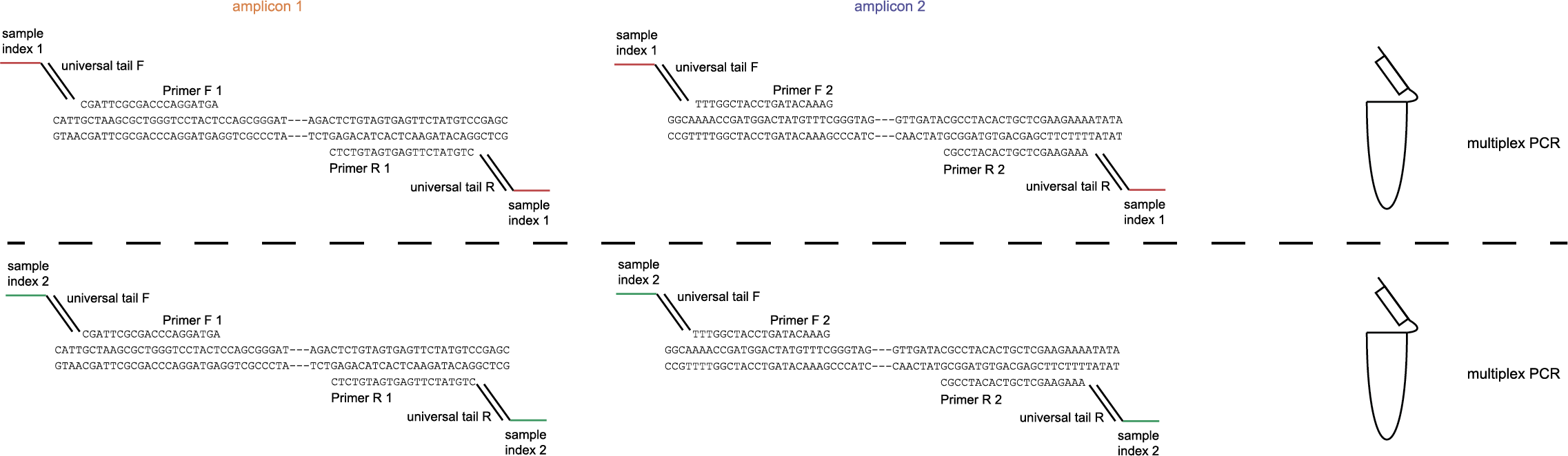
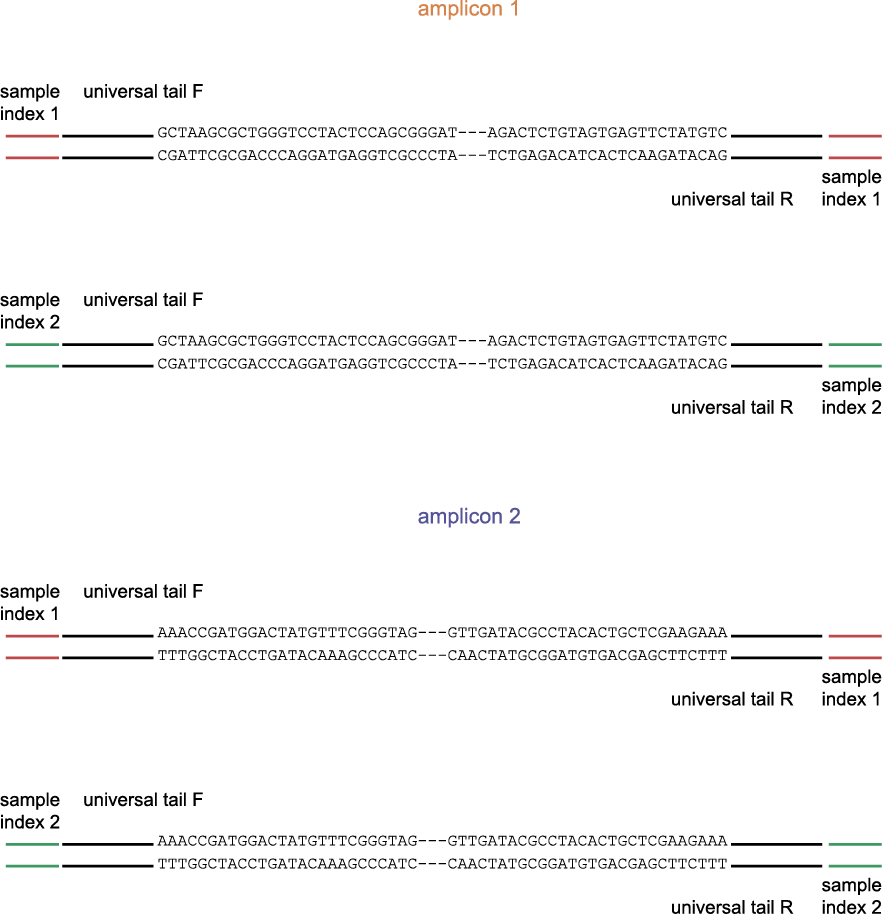
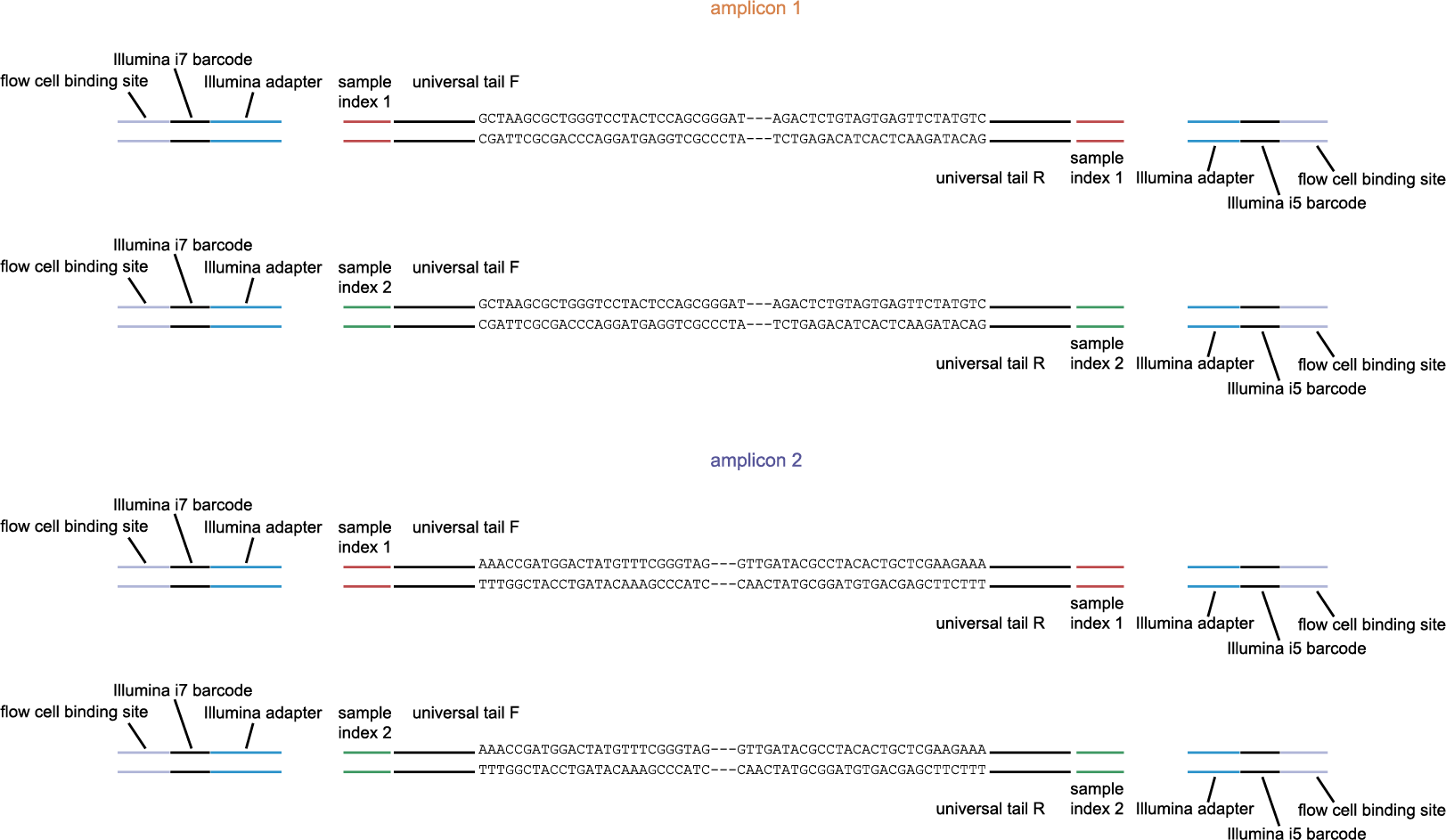

Recognizing haplotypes
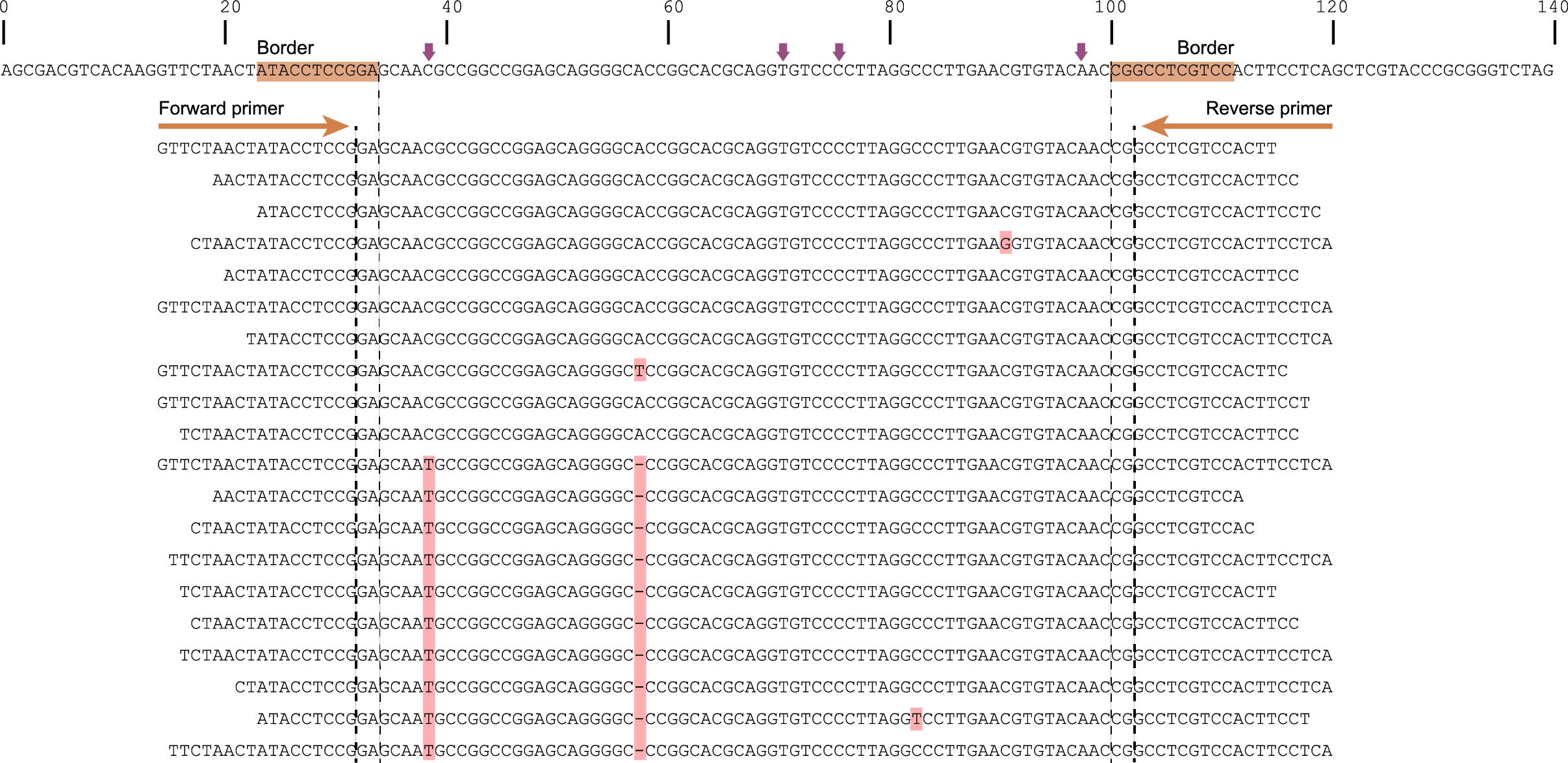
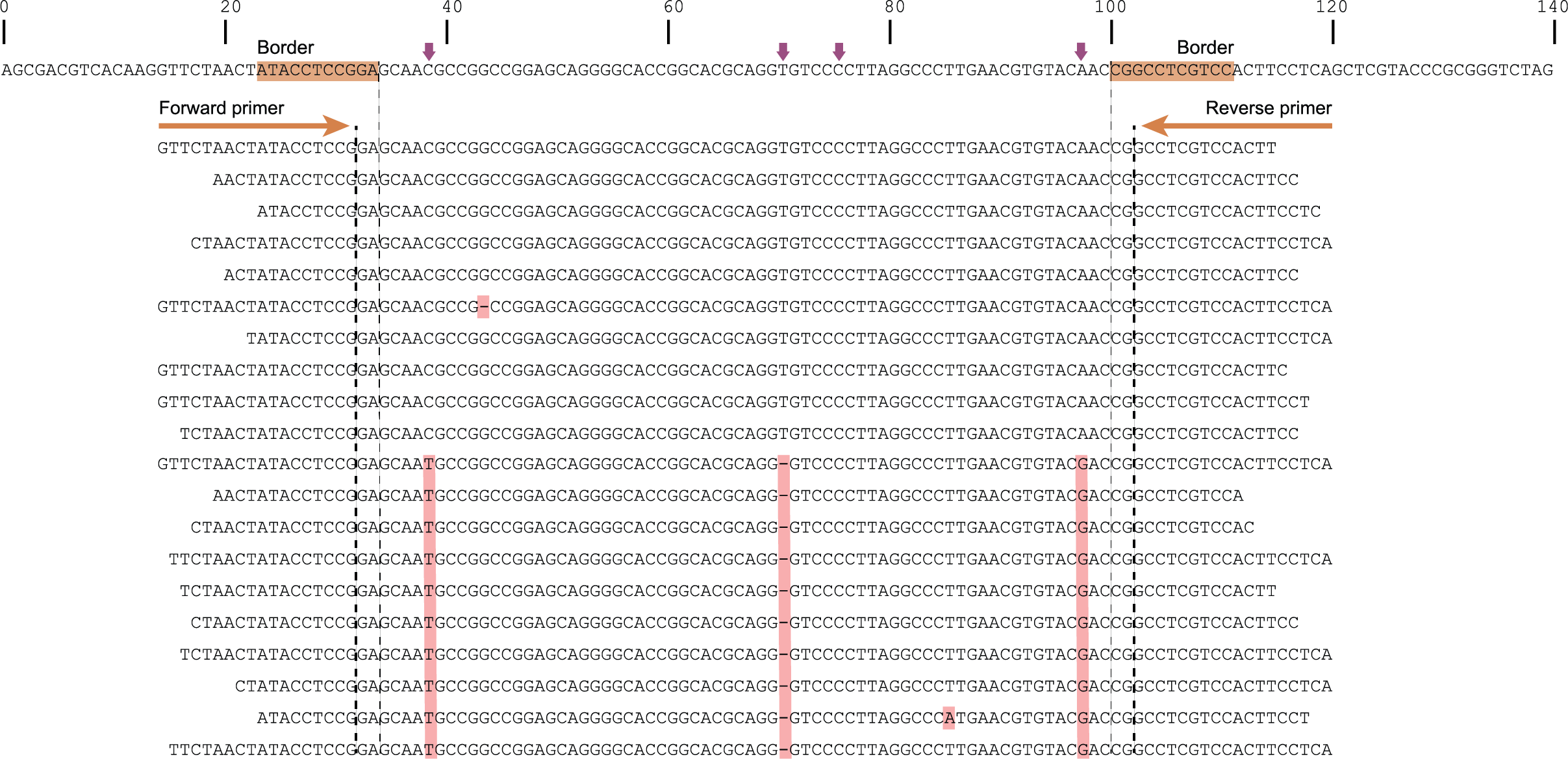
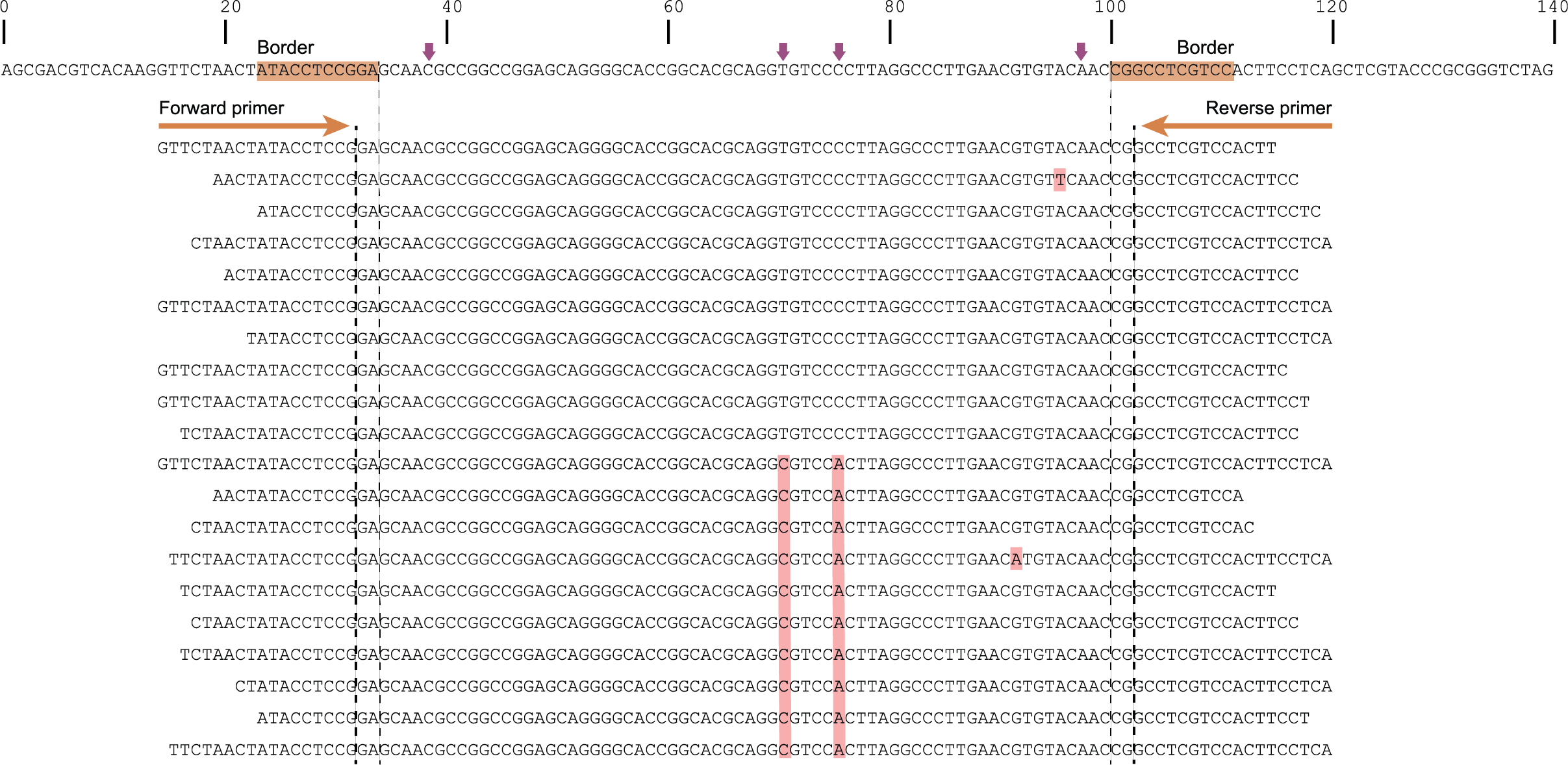
The tabs below show the same locus/amplicon in 3 diploid individuals. A total of 4 SNPs and 2 deletions are found among the individuals; 4 haplotypes can clearly be defined (1 reference allele and 3 alternative alleles).
Step 1: Extracting window-overlapping reads ID’s from BAM files and reads from FASTQ files
procedure
In order to run SMAP haplotype-window on HiPlex data, the user should create a custom GFF file with the desired Border positions enclosing Windows (see instructions here).
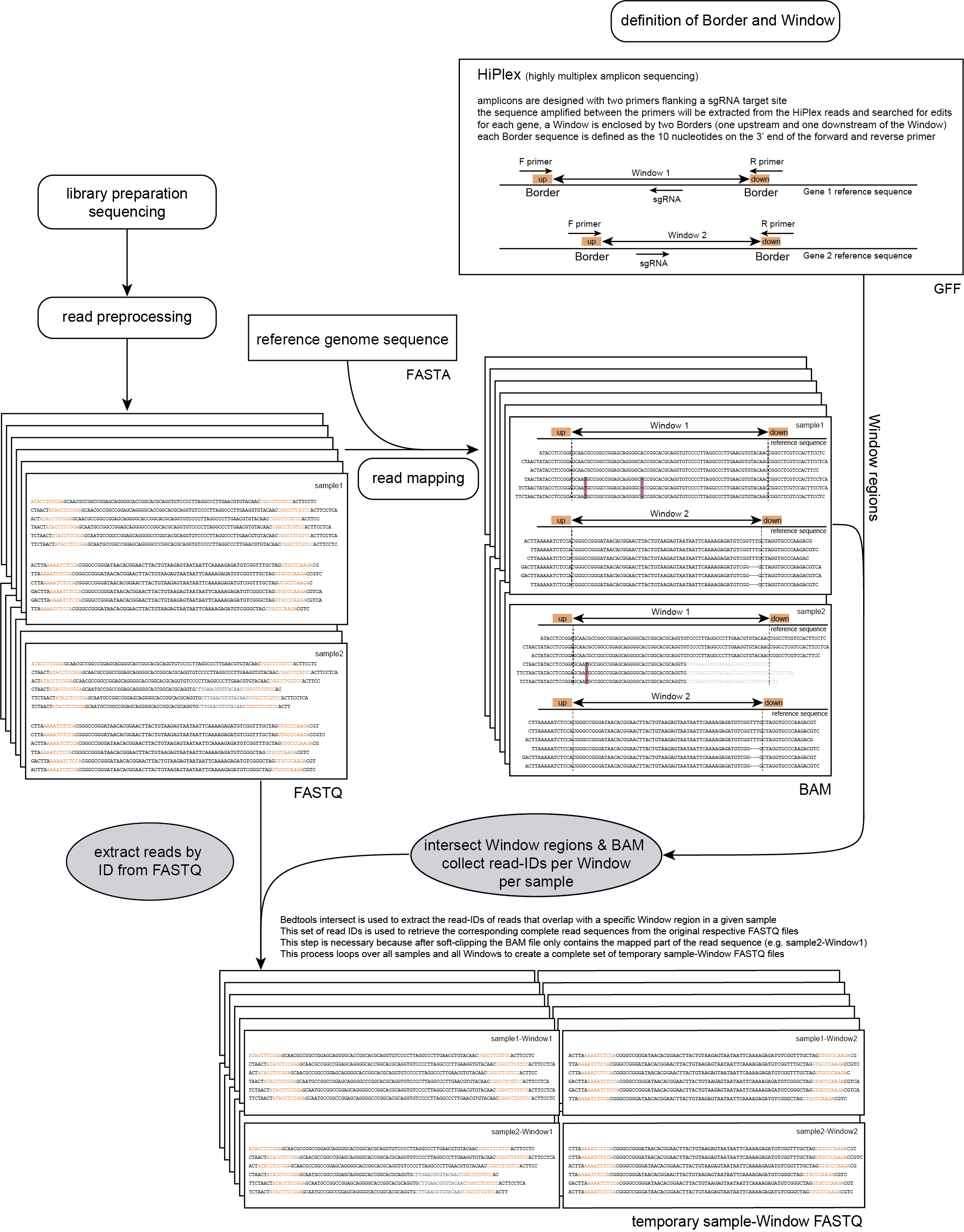
Step 2: Trimming and counting haplotypes
Per FASTQ file (one for each sample-Window combination), reads are passed to Cutadapt using the Window-specific pair of Border sequences for pattern trimming. Both Borders need to be found and trimmed, otherwise the read is discarded. This approach ensures the identification and removal of partial Window sequences, that would otherwise be mistaken for additional haplotypes. Because the Window is defined as the region inbetween the Borders (i.e. read regions retained after removal of the Borders), the entire read sequence spanning the Window is considered as a unique haplotype.
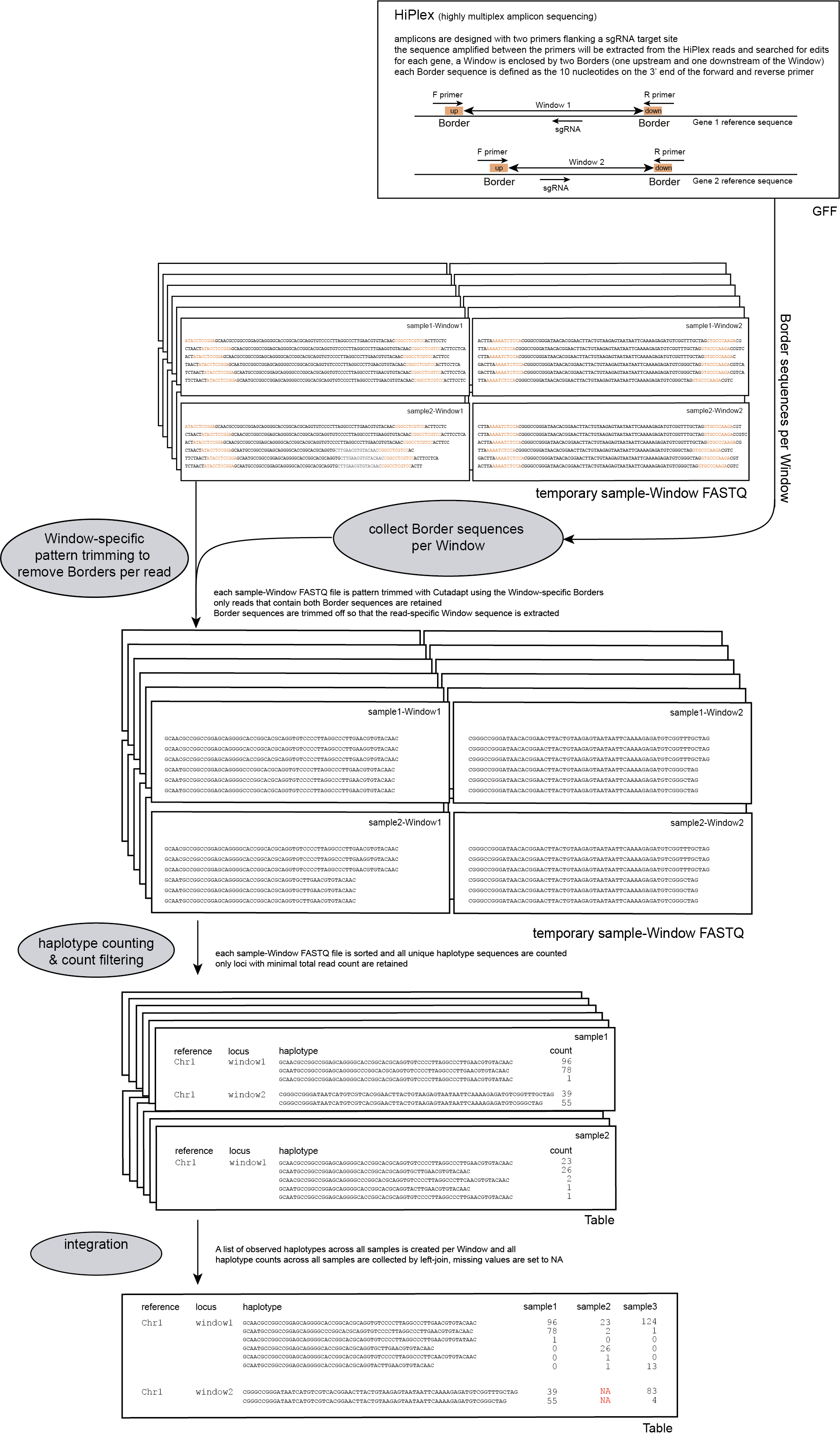
procedure
The following procedure is performed per sample:
-c, and the resulting haplotypes and counts are stored in tables.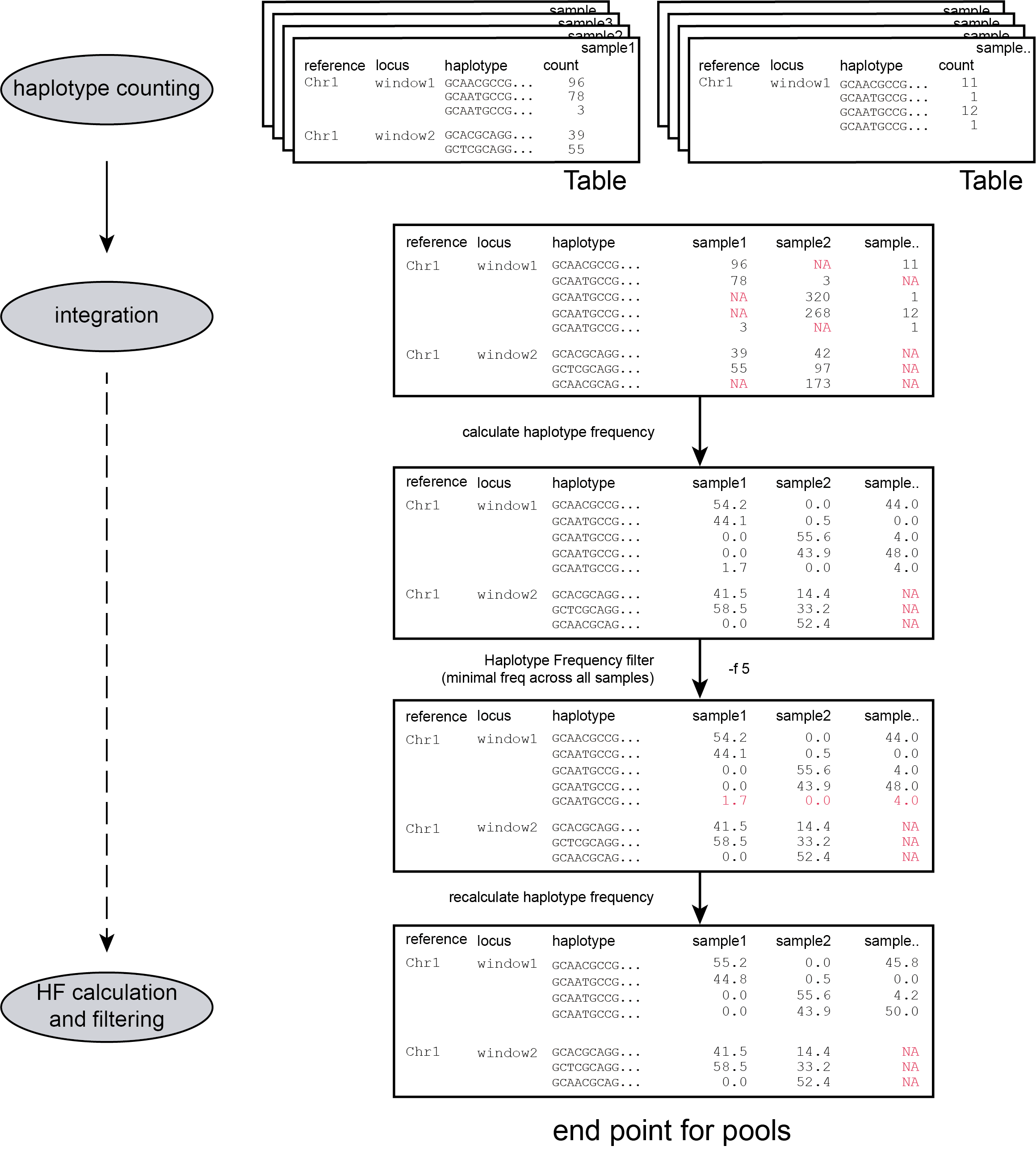
filters
loci with low read count are removed from the dataset with a read count threshold (option -c)
Accurate haplotype frequency estimation requires a minimum read count which is different between sample type (individuals and Pool-Seq) and ploidy levels.
The user is advised to use the read count threshold to ensure that the reported haplotype frequencies per locus are indeed based on sufficient read data. If a locus has a total haplotype count below the user-defined minimal read count threshold (option -c; default 0, recommended 10 for diploid individuals, 20 for tetraploid individuals, and 30 for pools) then all haplotype observations are removed for that sample. For more information see page Recommendations.
Only loci with an number of haplotypes between a custom interval across all samples are returned
-j, --min_distinct_haplotypes ### (int) ### Filter for the minimum number of distinct haplotypes per locus [0].
-k, --max_distinct_haplotypes ### (int) ### Filter for the maximum number of distinct haplotypes per locus [inf].
Only haplotypes with a relative frequency higher than a custom threshold in at least one sample are retained (see Step 3)
-f, --min_haplotype_frequency ### (int) ### Set minimal HF (in %) to retain the haplotype in the genotyping matrix. Haplotypes above this threshold in at least one of the FAST files are retained. Haplotypes that never reach this threshold in any of the FASTQ files are removed [0].