Pools
Step 3: Filtering haplotypes and calculating haplotype frequencies
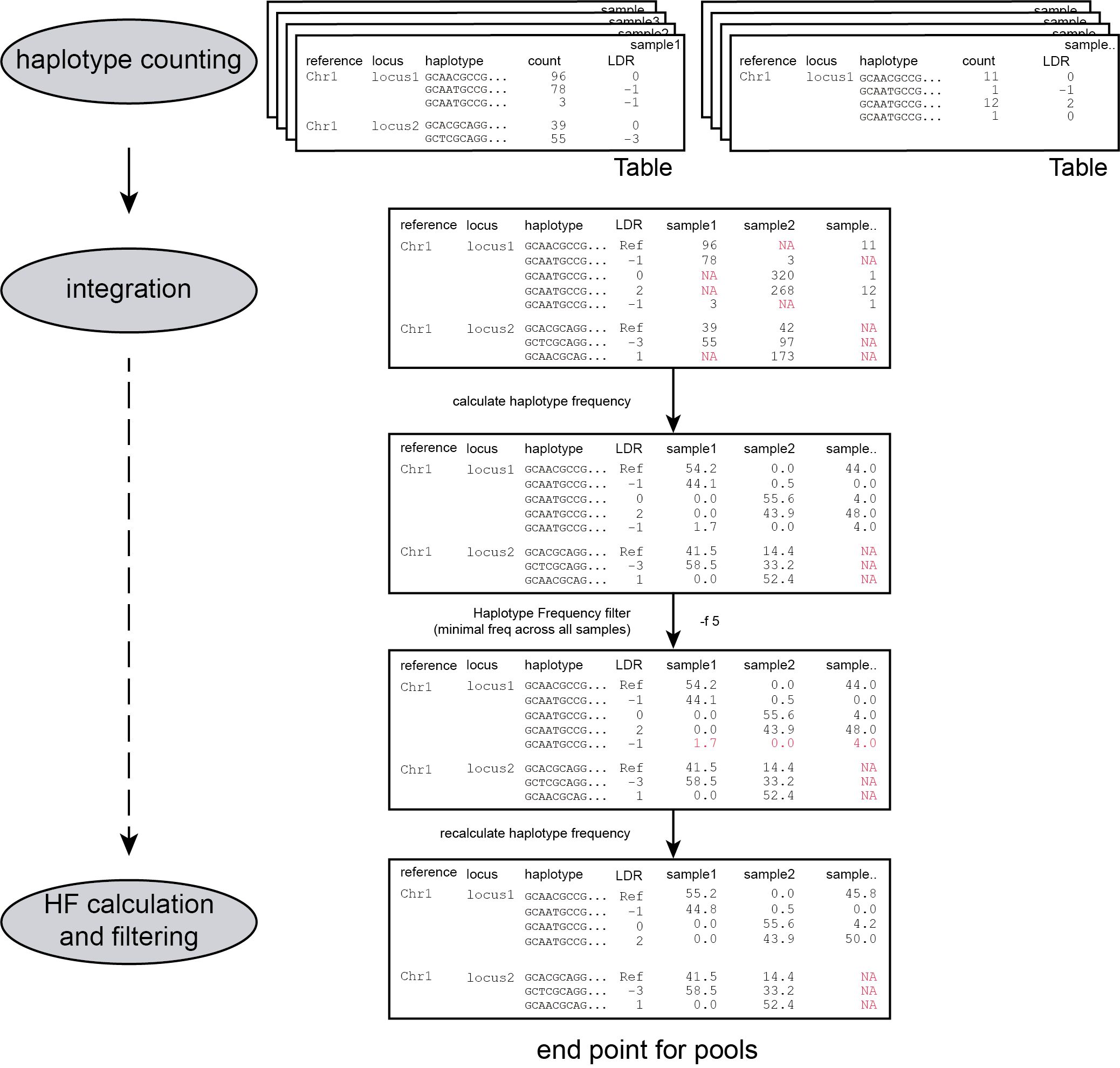
procedure
filters
read errors create false positive haplotypes
Randomly distributed read errors erroneously create low frequency haplotypes. Example data are shown in the scheme above, see tables below for the occurence in real HiPlex data of 8 diploid pools.
false positive haplotypes are removed from the data set with a minimal frequency threshold (option -f)
Haplotypes that never reach a user-defined minimal HF threshold (default 0%) in at least one of the BAM files, are removed entirely from the haplotype count table. After this filter, the haplotype frequency is recalculated on the remaining read counts per haplotype per Window per BAM file. The effect of adjusting this filter is illustrated in real data in the tables below. Please compare the tables below at subsequent steps of filtering: -f 0 filtering is effectively no filter (all haplotype observations > 0% are kept). Most noise from low frequency haplotypes can effectively be removed with -f 1 or -f 5 . We recommend to test the effect of this parameter on your own data, however in individuals it is best to have a high -f value, as alleles/haplotypes are expected to be present in around 25% and 50% of the reads in tetraploids and diploids respectively.
Samples with false positive haplotypes are masked for those haplotypes in the data set with a minimal frequency threshold (option -m)
Haplotypes that reach the user defined -f value in at least one BAM/FASTQ file are retained, however other BAM/FASTQ files that have a HF lower than the -f value for a haplotype where at least one sample exceeded the -f are also retained.
The option -m masks all HF lower than set value by substituting them with NaN’s or another value defined by --undefined_representation. The other haplotype frequencies are not recalculated.
The following tabs represent data from a real experiment. Three CRISPR/Cas9 targetted loci are shown. The number of haplotypes in -f 0 filtering is overwhelming, showcasing the importance of haplotype frequency filtering
# .. csv-table:: # :file: ../../.././tables/window/2n_ind_hiplex_r10_f0.csv # :header-rows: 1
# .. csv-table:: # :file: ../../.././tables/window/2n_ind_hiplex_r10_f1.csv # :header-rows: 1
# .. csv-table:: # :file: ../../.././tables/window/2n_ind_hiplex_r10_f5.csv # :header-rows: 1
# .. csv-table:: # :file: ../../.././tables/window/2n_ind_hiplex_r10_f5_m1.csv # :header-rows: 1
# .. csv-table:: # :file: ../../.././tables/window/2n_ind_hiplex_r10_f5_m5.csv # :header-rows: 1
After filtering out low frequency haplotypes the final haplotype frequency table is created. This is the end point for analysis of Pool-Seq data.
Haplotype frequency distributions
The different tabs below show the typical haplotype frequency distributions of HiPlex data in pools. The commands to run SMAP haplotype-window on these datatypes are shown below each graph.
# .. image:: ../../../images/window/tobemade
smap haplotype-window -alignments_dir /path/to/BAM/ -genome /path/to/RefGenome -borders /path/to/GFF -reads_dir /path/to/FASTQ -min_read_count 30 -f 2 -p 8 --min_distinct_haplotypes 2
# .. image:: ../../../images/window/tobemade
smap haplotype-window -alignments_dir /path/to/BAM/ -genome /path/to/RefGenome -borders /path/to/GFF -reads_dir /path/to/FASTQ -min_read_count 30 -f 2 -p 8 --min_distinct_haplotypes 2
Output
Tabular output
-c) and haplotype frequency (-f) filtered counts per haplotype per Window as defined in the GFF file.
Locus
Haplotypes
LDR
Sample1
Sample2
Sample..
Window_1
ACGTCGTCGC
ref
60
13
34
Window_1
ACGTCGTCAC
0
19
90
51
Window_2
GCTCATCG
ref
70
63
87
Window_2
GCTCTCG
-1
108
22
134
Locus
Haplotypes
LDR
Sample1
Sample2
Sample..
Window_1
ACGTCGTCGC
ref
0.76
0.13
0.40
Window_1
ACGTCGTCAC
0
0.24
0.87
0.60
Window_2
GCTCATCG
ref
0.39
0.74
0.39
Window_2
GCTCTCG
-1
0.61
0.26
0.61
-j (minimum distinct haplotypes) and -k (maximum distinct haplotypes), resulting in the file freqs_distinct_haplotypes_filter.tsvCode
-genome ################### (str) ### FASTA file with the reference genome sequence.–borders ################## (str) ### GFF file with the coordinates of pairs of Borders that enclose a Window. Must contain NAME=<> in column 9 to denote the Window name.–reads_dir ################# (str) ### Path to the directory containing FASTQ files with the reads mapped to the reference genome to create the BAM files. The FASTQ file names must have the same prefix as the BAM files specified in -alignments_dir [no default].-alignments_dir ############# (str) ### Path to the directory containing BAM and BAI alignment files. All BAM files should be in the same directory [no default].-–guides ################## (str) ### Optional FASTA file containing the sequences from sgRNAs used in CRISPR-Cas9 genome editing. Useful when amplicons on the CRISPR-Cas9/sgRNA delivery vector are included in the HiPlex amplicon mixture.-p, --processes ############ (int) ### Number of parallel processes [1].-o, --out ################ (str) ### Basename of the output file without extension [SMAP_haplotype_window].-u, --undefined_representation # (str) ### Value to use for non-existing or masked data [NaN].-h, --help ###################### Show the full list of options. Disregards all other parameters.-v, --version #################### Show the version. Disregards all other parameters.--debug ######################### Enable verbose logging. Provides additional intermediate output-files used for sample-specific QC.-q, --min_mapping_quality #### (int) ### Minimum .bam mapping quality for reads to be included in the analysis [30].-c, --min_read_count ####### (int) ### Minimal total number of reads per locus per sample [0].-d, --max_read_count ####### (int) ### Maximal number of reads per locus per sample, read depth is calculated after filtering out the low frequency haplotypes (-f) [inf].-f, --min_haplotype_frequency # (int) ### Set minimal haplotype frequency (in %) to retain the haplotype in the genotyping matrix. Haplotypes above this threshold in at least one of the samples are retained. Haplotypes that never reach this threshold in any of the samples are removed [0].-m, --mask_frequency ####### (float) ## Mask haplotype frequency values below this threshold for individual samples. Can be used to mask noise. Haplotypes are not removed based on this value, use --min_haplotype_frequency for this purpose instead.-j, --min_distinct_haplotypes # (int) ### Set minimal number of distinct haplotypes per locus across all samples. Loci that do not fit this criteria are removed from the final output [0].-k, --max_distinct_haplotypes # (int) ### Set maximal number of distinct haplotypes per locus across all samples. Loci that do not fit this criteria are removed from the final output [inf].